








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (8): 284-293.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1467
Previous Articles Next Articles
LIU Na( ), LIU Shi-ke, WANG Qian-nan(
), LIU Shi-ke, WANG Qian-nan( )
)
Received:2020-12-01
Online:2021-08-26
Published:2021-09-10
Contact:
WANG Qian-nan
E-mail:1032047781@qq.com;wangqiannan@hainanu.edu.cn
LIU Na, LIU Shi-ke, WANG Qian-nan. Construction of a Strain with Fluorescence Labeling of Cytoskeleton in Colletotrichum gloeosporioides[J]. Biotechnology Bulletin, 2021, 37(8): 284-293.

Fig. 1 Diagram of fluorescent labeled cytoskeleton vectors A: The diagram of microfilament labeling vector through Lifeact-EGFP; LA: coding sequence of Lifeact; PgpdA: promoter of glyceraldehyde-3-phosphate dehydrogenase in Aspergillus nidulans; TtrpC: terminator of tryptophan synthesis gene in Aspergillus nidulans; HPT:Hygromycin phosphate transferase resistance gene. B: The diagram of microtubule labeling vector through MBD-EGFP; MBD:microtubule binding domain. C: The diagram of inserting EGFP coding sequence behind gene CgTUB1 through homologous recombination; WT: wild type; Mutant: mutant strain; Flanking region: downstream of gene CgTUB1
| 引物名称Primer name | 序列Primer sequences(5'-3') | 用途Purpose | 内切酶位点Restriction enzyme |
|---|---|---|---|
| Lifeact-EGFP-F | tctagaATGGGTGTCGCAGATTTGA | Lifeact-EGFP扩增 Amplification for Lifeact-EGFP | Xba I |
| EGFP-R | gagctcTTACTTGTACAGCTCGTCC | Lifeact-EGFP扩增 Amplification for Lifeact-EGFP | Sac I |
| MBD-EGFP-F | tctagaATGAGCCTCGCCTCAGG | MBD-EGFP扩增 Amplification for MBD-EGFP | Xba I |
| EGFP-R | gagctcTTACTTGTACAGCTCGTCC | MBD-EGFP扩增 Amplification for MBD-EGFP | Sac I |
| CgTub-SF | tctagaAGCCCTACAACGCCACT | CgTUB1 DNA 3'端扩增 Amplification for CgTUB1 DNA 3' | Xba I |
| CgTub-R | ggtaccAACCTCCTCCTCAAGAGG | CgTUB1 DNA 3'端扩增 Amplification for CgTUB1 DNA 3' | Kpn I |
| hph-SPLR | GGATGCCTCCGCTCGAAGTA | HPT-5'端扩增 Amplification for HPT-5' | - |
| hph-SPLF | CGTTGCAAGACCTGCCTGAA | HPT-3'端扩增 Amplification for HPT-3' | - |
| CgTub-MR2 | GCAGTTATTAGACTGCGCAACTGG- TTCCCGGTCGG | HPT-3'端扩增 Amplification for HPT-3' | - |
| CgTub-MF2 | CCGACCGGGAACCAGTTGCGCAGT- CTAATAACTGC | CgTUB1下游序列扩增 Amplification for CgTUB1 downstream fragment | - |
| CgTub-3R | TGCCTATACCGAACACGATC | CgTUB1下游序列扩增 Amplification for CgTUB1 downstream fragment | - |
| CgTubegfp-JCF | ACTGACCTTCGCTCCTACCCA | 敲入重组菌株检测 PCR diagnosis of the CgTUB1-EGFP labeling strains | - |
| CgTubegfp-JCR | TGCCGTTCTTCTGCTTGTCG | 敲入重组菌株检测 PCR diagnosis of the CgTUB1-EGFP labeling strains | - |
| HPT-JCF | ACAGCGGTCATTGACTGGAGCGA | 敲入重组菌株检测 PCR diagnosis of the CgTUB1-EGFP labeling strains | - |
| CgTub-JC3R | GCCCTTTGGAATACCCATCTC | 敲入重组菌株检测 PCR diagnosis of the CgTUB1-EGFP labeling strains | - |
Table 1 Primers used in this study
| 引物名称Primer name | 序列Primer sequences(5'-3') | 用途Purpose | 内切酶位点Restriction enzyme |
|---|---|---|---|
| Lifeact-EGFP-F | tctagaATGGGTGTCGCAGATTTGA | Lifeact-EGFP扩增 Amplification for Lifeact-EGFP | Xba I |
| EGFP-R | gagctcTTACTTGTACAGCTCGTCC | Lifeact-EGFP扩增 Amplification for Lifeact-EGFP | Sac I |
| MBD-EGFP-F | tctagaATGAGCCTCGCCTCAGG | MBD-EGFP扩增 Amplification for MBD-EGFP | Xba I |
| EGFP-R | gagctcTTACTTGTACAGCTCGTCC | MBD-EGFP扩增 Amplification for MBD-EGFP | Sac I |
| CgTub-SF | tctagaAGCCCTACAACGCCACT | CgTUB1 DNA 3'端扩增 Amplification for CgTUB1 DNA 3' | Xba I |
| CgTub-R | ggtaccAACCTCCTCCTCAAGAGG | CgTUB1 DNA 3'端扩增 Amplification for CgTUB1 DNA 3' | Kpn I |
| hph-SPLR | GGATGCCTCCGCTCGAAGTA | HPT-5'端扩增 Amplification for HPT-5' | - |
| hph-SPLF | CGTTGCAAGACCTGCCTGAA | HPT-3'端扩增 Amplification for HPT-3' | - |
| CgTub-MR2 | GCAGTTATTAGACTGCGCAACTGG- TTCCCGGTCGG | HPT-3'端扩增 Amplification for HPT-3' | - |
| CgTub-MF2 | CCGACCGGGAACCAGTTGCGCAGT- CTAATAACTGC | CgTUB1下游序列扩增 Amplification for CgTUB1 downstream fragment | - |
| CgTub-3R | TGCCTATACCGAACACGATC | CgTUB1下游序列扩增 Amplification for CgTUB1 downstream fragment | - |
| CgTubegfp-JCF | ACTGACCTTCGCTCCTACCCA | 敲入重组菌株检测 PCR diagnosis of the CgTUB1-EGFP labeling strains | - |
| CgTubegfp-JCR | TGCCGTTCTTCTGCTTGTCG | 敲入重组菌株检测 PCR diagnosis of the CgTUB1-EGFP labeling strains | - |
| HPT-JCF | ACAGCGGTCATTGACTGGAGCGA | 敲入重组菌株检测 PCR diagnosis of the CgTUB1-EGFP labeling strains | - |
| CgTub-JC3R | GCCCTTTGGAATACCCATCTC | 敲入重组菌株检测 PCR diagnosis of the CgTUB1-EGFP labeling strains | - |

Fig. 2 PCR diagnosis of cytoskeleton labeling recombinant strains A: PCR diagnosis of microfilament labeling strains by Lifeact-EGFP. B:PCR diagnosis of microtubule labeling strains by MBD-EGFP. C: PCR diagnosis of the upstream recombinant fragment of CgTUB1-EGFP labeling strains. D: PCR diagnosis of the downstream recombinant fragment of CgTUB1-EGFP labeling strains. M: DNA marker DM5000

Fig. 3 Cytoskeleton structure in conidiophores of fluorescent labeled recombinant strains CK: Control strain expressing EGFP; Lifeact-EGFP: microfilament labeled recombinant strain; CgTUB1-EGFP: microtubule labeled recombinant strain; Z Projection:stacked image of Z-series slices; scale bar:5 μm. The same below

Fig. 5 Clony diameters and conidiations of WT and mutant strains A: Clony diameter of WT and mutant strains on PDA media; B: conidiation of WT and mutant strains on PDB liquid media; WT: wild type strain
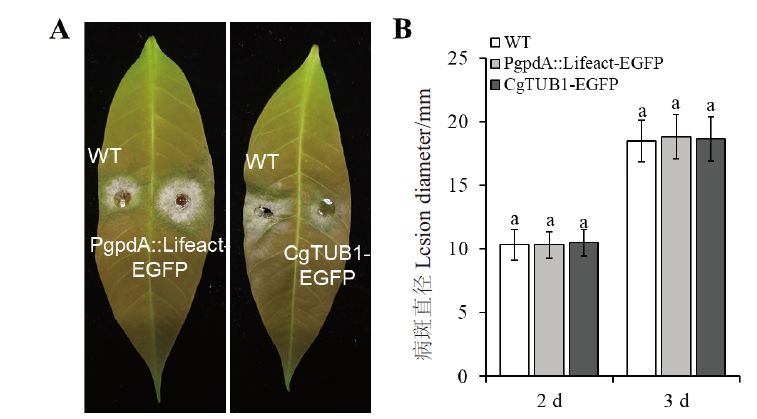
Fig. 6 Virulence assay on rubber tree leaves and lesion diameter after inoculation with WT and recom-binant strains A: Disease symptoms of rubber tree leaves at day 3 after inoculation (dpi); B: Mean lesion diameters of rubber tree leaves at day 2 and 3 (dpi).
| [1] | Riquelme M, Aguirre J, Bartnicki-García S, et al. Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures[J]. Microbiology and Molecular Biology Reviews, 2018, 82(2):e00068-17. |
| [2] |
Riquelme M. Tip growth in filamentous fungi:a road trip to the apex[J]. Annual Review of Microbiology, 2013, 67(1):587-609.
doi: 10.1146/annurev-micro-092412-155652 URL |
| [3] | 郭云峰, 安邦. 橡胶树胶孢炭疽菌NADPH氧化酶功能研究[J]. 生物技术通报, 2018, 34(10):165-171. |
| Guo YF, An B. Functional analysis of NADPH oxidases in Colletotrichum gloeosporioides from Hevea brasiliensis[J]. Biotechnology Bulletin, 2018, 34(10):165-171. | |
| [4] |
Bartnicki-Garcia S, Bartnicki DD, Gierz G, et al. Evidence that spitzenkörper behavior determines the shape of a fungal hypha:a test of the hyphoid model[J]. Experimental Mycology, 1995, 19(2):153-159.
pmid: 7614375 |
| [5] |
Howard RJ, Aist JR. Cytoplasmic microtubules and fungal morphogenesis:ultrastructural effects of methyl Benzimidazole-2-Ylcarbamate determined by freeze-substitution of hyphal tip cells[J]. The Journal of Cell Biology, 1980, 87(1):55-64.
doi: 10.1083/jcb.87.1.55 URL |
| [6] |
Read ND. Exocytosis and growth do not occur only at hyphal tips[J]. Molecular Microbiology, 2011, 81(1):4-7.
doi: 10.1111/mmi.2011.81.issue-1 URL |
| [7] |
Taheri-Talesh N, Horio T, Araujo-Bazán L, et al. The tip growth apparatus of Aspergillus nidulans[J]. Molecular Biology of the Cell, 2008, 19(4):1439-1449.
doi: 10.1091/mbc.E07-05-0464 pmid: 18216285 |
| [8] |
Sudbery P. Fluorescent proteins illuminate the structure and function of the hyphal tip apparatus[J]. Fungal Genetics and Biology, 2011, 48(9):849-857.
doi: 10.1016/j.fgb.2011.02.004 pmid: 21362491 |
| [9] |
Lappalainen P. Actin-binding proteins:the long road to understanding the dynamic landscape of cellular actin networks[J]. Molecular Biology of the Cell, 2016, 27(16):2519-2522.
doi: 10.1091/mbc.E15-10-0728 pmid: 27528696 |
| [10] |
Pollard TD. Actin and actin-binding proteins[J]. Cold Spring Harbor Perspectives in Biology, 2016, 8(8):a018226.
doi: 10.1101/cshperspect.a018226 URL |
| [11] |
Riedl J, Crevenna AH, Kai K, et al. Lifeact:a versatile marker to visualize F-actin[J]. Nature Methods, 2008, 5(7):605.
doi: 10.1038/nmeth.1220 URL |
| [12] | 王倩男, 张晓东, 罗红丽, 等. 巴西橡胶树叶肉细胞原生质体中细胞骨架结构及动态观察[J]. 热带作物学报, 2018, 39(3):494-501. |
| Wang QN, Zhang XD, Lou HL, et al. Visualization of cytoskeleton organization and dynamics in the rubber tree(Hevea brasiliensis)mesophyll protoplasts[J]. Chinese Journal of Tropical Crops, 2018, 39(3):494-501. | |
| [13] |
Li YB, Xu R, Liu C, et al. Magnaporthe oryzae fimbrin organizes actin networks in the hyphal tip during polar growth and pathogenesis[J]. PLoS Pathogens, 2020, 16(3):e1008437.
doi: 10.1371/journal.ppat.1008437 URL |
| [14] |
Schumacher J. Tools for Botrytis cinerea:New expression vectors make the gray mold fungus more accessible to cell biology approaches[J]. Fungal Genetics and Biology, 2012, 49(6):483-497.
doi: 10.1016/j.fgb.2012.03.005 pmid: 22503771 |
| [15] |
Marc J, Granger CL, Brincat J, et al. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells[J]. The Plant Cell, 1998, 10(11):1927-1940.
pmid: 9811799 |
| [16] |
Wang QN, An B, Hou XR, et al. Dicer-like proteins regulate the growth, conidiation, and pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis[J]. Frontiers in Microbiology, 2018, 8:2621.
doi: 10.3389/fmicb.2017.02621 URL |
| [17] |
Harris SD, Momany M. Polarity in filamentous fungi:moving beyond the yeast paradigm[J]. Fungal Genetics and Biology, 2004, 41(4):391-400.
doi: 10.1016/j.fgb.2003.11.007 URL |
| [18] |
Berepiki A, Lichius A, Read ND. Actin organization and dynamics in filamentous fungi[J]. Nature Reviews Microbiology, 2011, 9(12):876-887.
doi: 10.1038/nrmicro2666 pmid: 22048737 |
| [19] |
Li LW, Chen XL, Zhang SP, et al. MoCAP proteins regulated by MoArk1-mediated phosphorylation coordinate endocytosis and actin dynamics to govern development and virulence of Magnaporthe oryzae[J]. PLoS Genetics, 2017, 13(5):e1006814.
doi: 10.1371/journal.pgen.1006814 URL |
| [20] |
Li X, Gao C, Li L, et al. MoEnd3 regulates appressorium formation and virulence through mediating endocytosis in rice blast fungus Magnaporthe oryzae[J]. PLoS Pathogens, 2017, 13(6):e1006449.
doi: 10.1371/journal.ppat.1006449 URL |
| [21] |
Liu, ZY, Wu SS, Chen Y, et al. The microtubule end-binding protein FgEB1 regulates polar growth and fungicide sensitivity via different interactors in Fusarium graminearum[J]. Environmental Microbiology, 2017, 19(5):1791-1807.
doi: 10.1111/emi.2017.19.issue-5 URL |
| [22] |
Aizawa H, Sameshima M, Yahara I. A green fluorescent protein-actin fusion protein dominantly inhibits cytokinesis, cell spreading, and locomotion in Dictyostelium[J]. Cell Structure and Function, 1997, 22:335-345.
pmid: 9248997 |
| [23] |
Wang YS, Motes CM, Mohamalawari DR, et al. Green fluorescent protein fusions to Arabidopsis fimbrin 1 for spatio-temporal imaging of F-actin dynamics in roots[J]. Cell Motility and the Cytoskeleton, 2004, 59(2):79-93.
doi: 10.1002/cm.v59:2 URL |
| [24] |
Riedl J, Crevenna AH, Kai K, et al. Lifeact:a versatile marker to visualize F-actin[J]. Nature Methods, 2008, 5(7):605-607.
doi: 10.1038/nmeth.1220 URL |
| [25] | Riedl J, Flynn Kevin C, Raducanu A, et al. Lifeact mice for studying F-actin dynamics[J]. Nature Methods, 2010, (3):168-169. |
| [26] |
Era A, Tominaga M, Ebine K, et al. Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha[J]. Plant and Cell Physiology, 2009, 50(6):1041-1048.
doi: 10.1093/pcp/pcp055 URL |
| [27] |
Vidali L, Rounds CM, Hepler PK, et al. Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells[J]. PLoS One, 2009, 4(5):e5744.
doi: 10.1371/journal.pone.0005744 URL |
| [28] |
Smertenko AP, Deeks MJ, Hussey PJ. Strategies of actin reorganisation in plant cells[J]. Journal of Cell Science, 2010, 123(17):3019-3028.
doi: 10.1242/jcs.071126 URL |
| [29] |
Qu XL, Zhang H, Xie YR, et al. Arabidopsis villins promote actin turnover at pollen tube tips and facilitate the construction of actin collars[J]. The Plant Cell, 2013, 25(5):1803-1817.
doi: 10.1105/tpc.113.110940 URL |
| [30] |
Delgado-Alvarez DL, Callejas-Negrete OA, Gómez N, et al. Visualization of F-actin localization and dynamics with live cell markers in Neurospora crassa[J]. Fungal Genetics and Biology, 2010, 47(7):573-586.
doi: 10.1016/j.fgb.2010.03.004 pmid: 20302965 |
| [31] |
Ueda EI, Kashiwazaki J, Inoué S, et al. Fission yeast adf1 is necessary for reassembly of actin filaments into the contractile ring during cytokinesis[J]. Biochemical and Biophysical Research Communications, 2018, 506(2):330-338.
doi: 10.1016/j.bbrc.2018.07.156 URL |
| [32] |
Christensen JR, Homa KE, Morganthaler AN, et al. Cooperation between tropomyosin and α-actinin inhibits fimbrin association with actin filament networks in fission yeast[J]. eLife, 2019, 8:e47279.
doi: 10.7554/eLife.47279 URL |
| [33] | 陈斌, 田娟, 冯志迪, 等. 大丽轮枝菌微丝荧光标记载体构建及应用[J]. 生物工程学报, 2019, 35(8):1520-1528. |
| Chen B, Tian J, Feng ZD, et al. Construction and application of actin fluorescent marker in Verticillium dahliae Kleb[J]. Chinese Journal of Biotechnology, 2019, 35(8):1520-1528. | |
| [34] |
Araujo-Bazán L, Penalva MA, Espeso EA. Preferential localization of the endocytic internalization machinery to hyphal tips underlies polarization of the actin cytoskeleton in in Aspergillus nidulans[J]. Molecular Microbiology, 2008, 67:891-905.
doi: 10.1111/j.1365-2958.2007.06102.x pmid: 18179595 |
| [35] |
Huang L, Zhang S, Yin Z, et al. MoVrp1, a putative verprolin protein, is required for asexual development and infection in the rice blast fungus Magnaporthe oryzae[J]. Scientific Reports, 2017, 7:41148.
doi: 10.1038/srep41148 pmid: 28117435 |
| [36] |
Wang Q, Huang S. Visualization of microtubule organization and dynamics in living Arabidopsis embryonic cells[J]. Molecular Plant, 2014, 7(8):1397-1401.
doi: 10.1093/mp/ssu038 URL |
| [37] |
Chan J, Calder G, Fox S, et al. Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells[J]. Nature Cell Biology, 2007, 9(2):171-175.
doi: 10.1038/ncb1533 URL |
| [38] |
Konzack S, Rischitor PE, Enke C, et al. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans[J]. Molecular Biology of the Cell, 2005, 16(2):497-506.
pmid: 15563609 |
| [39] |
Zeng CJ, Kim HR, Vargas Arispuro I, et al. Microtubule plus end-tracking proteins play critical roles in directional growth of hyphae by regulating the dynamics of cytoplasmic microtubules in Aspergillus nidulans[J]. Molecular Microbiology, 2014, 94(3):506-521.
doi: 10.1111/mmi.2014.94.issue-3 URL |
| [40] |
Ramos-Garcia SL, Roberson RW, Freitag M, et al. Cytoplasmic bulk flow propels nuclei in mature hyphae of Neurospora crassa[J]. Eukaryot Cell, 2009, 8(12):1880-1890.
doi: 10.1128/EC.00062-09 pmid: 19684281 |
| [41] |
Luo YP, Zhang HC, Qi LL, et al. FgKin1 kinase localizes to the septal pore and plays a role in hyphal growth, ascospore germination, pathogenesis, and localization of Tub1 beta-tubulins in Fusarium graminearum[J]. The New Phytologist, 2014, 204(4):943-954.
doi: 10.1111/nph.2014.204.issue-4 URL |
| [42] |
Zhou Y, Zhu Y, Li Y, et al. β1 Tubulin rather than β2 tubulin is the preferred binding target for carbendazim in Fusarium graminearum[J]. Phytopathology, 2016, 106(9):978-985.
doi: 10.1094/PHYTO-09-15-0235-R URL |
| [43] |
Takeshita N, Manck R, Grün N, et al. Interdependence of the actin and the microtubule cytoskeleton during fungal growth[J]. Current Opinion in Microbiology, 2014, 20:34-41.
doi: 10.1016/j.mib.2014.04.005 pmid: 24879477 |
| [1] | ZHANG Le-le, WANG Guan, LIU Feng, HU Han-qiao, REN Lei. Isolation, Identification and Biocontrol Mechanism of an Antagonistic Bacterium Against Anthracnose on Mango Caused by Colletotrichum gloeosporioides [J]. Biotechnology Bulletin, 2023, 39(4): 277-287. |
| [2] | LIU Sha-yu, CAO Jian, LI Meng, LIU Zhi-qiang, LI Xiao-yu. Biological Function of a Zn2Cys6 Transcription Factor CgAswA in Colletotrichum gloeosporioides [J]. Biotechnology Bulletin, 2021, 37(9): 161-170. |
| [3] | GUO Yun-feng, AN Bang. Functional Analysis of NADPH Oxidases in Colletotrichum gloeosporioides from Hevea brasiliensis [J]. Biotechnology Bulletin, 2018, 34(10): 165-171. |
| [4] | SU Chu-lian, KANG Hao, MEI Zhi-dong, YANG Shi-you, LIU Xiao-mei, PU Jin-ji, ZHANG He. Expression Analysis of Pathogenic Genes During the Infection of Colletotrichum gloeosporioides to Mango [J]. Biotechnology Bulletin, 2018, 34(10): 182-186. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||