








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (11): 197-211.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0590
Previous Articles Next Articles
LIU Xiao-han( ), LIN Zi-xin, XIU Yu, DANG Yuan, LIN Shan-zhi(
), LIN Zi-xin, XIU Yu, DANG Yuan, LIN Shan-zhi( )
)
Received:2021-05-04
Online:2021-11-26
Published:2021-12-03
Contact:
LIN Shan-zhi
E-mail:han151294@163.com;linsz2002@163.com
LIU Xiao-han, LIN Zi-xin, XIU Yu, DANG Yuan, LIN Shan-zhi. Analysis of the MATE Family in the Seeds of Prunus sibirica and Cloning and Expression of Its Important Member MATE40[J]. Biotechnology Bulletin, 2021, 37(11): 197-211.
| 基因Gene | 引物序列Primer sequence(5'-3') | 基因Gene | 引物序列Primer sequence(5'-3') | |
|---|---|---|---|---|
| PsMATE14 | F:GAAGTGAAGAGGTTGGGATACATAG | PsMATE40 | F:AACAATGGCATCCAAGTTTTCGCCT | |
| R:CAATCGCAGTGCTGGAAA | R:GGAGCACACCAGTCAAAGTTAAGAG | |||
| PsMATE16A | F:ATGGATGCCGAAGAACAA | PsMATE41A | F:GAAGACACACTAAACAACCCTGAGC | |
| R:GCTGCTTCTTCACTTCCTTGATAAT | R:TGAGCAAGTAAACCACCA | |||
| PsMATE16B | F:AAGAAGTAAAGAAGCAGC | PsMATE41B | F:ATGGCGATGGCTTGGAGT | |
| R:TTCACCAAGATGACCTAC | R:GGCGAAAGAGGAGATTGAGT | |||
| PsMATE23 | F:TGGAGGGAGACCAAGACG | PsMATE41C | F:CAAGGCTGCTCTGGATTT | |
| R:GAGCCAATGTGACCAATGAA | R:GCTCCAAGGCACTCAAAT | |||
| PsMATE27 | F:TTCAACCGTCCAATCAAA | PsMATE48 | F:CTTCCTCATTTCTCCCTT | |
| R:GGAATAAGACGCAACACG | R:TGGCCTTGATTTCTTGTA | |||
| PsMATE32 | F:ATGGAGGATAAGAAGCAGC | PsMATE49 | F:TGCTCTACTCACGCACAA | |
| R:GGAAATGAAAGAAGGGTGT | R:AATAGCAAGCGAACCACC | |||
| PsMATE33 | F:CCTTGGGTGCTCTCACTCAGGTCTA | PsMATE51 | F:TCCTTCTGCTGCTTCATC | |
| R:TGACACCGAAAGCGAGGC | R:TAATGGCGGTGTTTGGTA | |||
| PsMATE35A | F:TATGTTTTGGATTGAGACAGTGAAG | PsMATE56 | F:TGCCAAGATAGCAACCAC | |
| R:TTCCGAGATGACCAACAAAGAG | R:CACCACAGAGCCCATTTA | |||
| PsMATE35B | F:GGTCGGTGACTTGGATAGA | UBC | F:GAGACCAGCAATAACCGTGAA | |
| R:TGAAGGTTTCCGAGGTGT | R:TCTTGTACTCCGTGGCATCCT |
Table 1 Primers used for qRT-PCR
| 基因Gene | 引物序列Primer sequence(5'-3') | 基因Gene | 引物序列Primer sequence(5'-3') | |
|---|---|---|---|---|
| PsMATE14 | F:GAAGTGAAGAGGTTGGGATACATAG | PsMATE40 | F:AACAATGGCATCCAAGTTTTCGCCT | |
| R:CAATCGCAGTGCTGGAAA | R:GGAGCACACCAGTCAAAGTTAAGAG | |||
| PsMATE16A | F:ATGGATGCCGAAGAACAA | PsMATE41A | F:GAAGACACACTAAACAACCCTGAGC | |
| R:GCTGCTTCTTCACTTCCTTGATAAT | R:TGAGCAAGTAAACCACCA | |||
| PsMATE16B | F:AAGAAGTAAAGAAGCAGC | PsMATE41B | F:ATGGCGATGGCTTGGAGT | |
| R:TTCACCAAGATGACCTAC | R:GGCGAAAGAGGAGATTGAGT | |||
| PsMATE23 | F:TGGAGGGAGACCAAGACG | PsMATE41C | F:CAAGGCTGCTCTGGATTT | |
| R:GAGCCAATGTGACCAATGAA | R:GCTCCAAGGCACTCAAAT | |||
| PsMATE27 | F:TTCAACCGTCCAATCAAA | PsMATE48 | F:CTTCCTCATTTCTCCCTT | |
| R:GGAATAAGACGCAACACG | R:TGGCCTTGATTTCTTGTA | |||
| PsMATE32 | F:ATGGAGGATAAGAAGCAGC | PsMATE49 | F:TGCTCTACTCACGCACAA | |
| R:GGAAATGAAAGAAGGGTGT | R:AATAGCAAGCGAACCACC | |||
| PsMATE33 | F:CCTTGGGTGCTCTCACTCAGGTCTA | PsMATE51 | F:TCCTTCTGCTGCTTCATC | |
| R:TGACACCGAAAGCGAGGC | R:TAATGGCGGTGTTTGGTA | |||
| PsMATE35A | F:TATGTTTTGGATTGAGACAGTGAAG | PsMATE56 | F:TGCCAAGATAGCAACCAC | |
| R:TTCCGAGATGACCAACAAAGAG | R:CACCACAGAGCCCATTTA | |||
| PsMATE35B | F:GGTCGGTGACTTGGATAGA | UBC | F:GAGACCAGCAATAACCGTGAA | |
| R:TGAAGGTTTCCGAGGTGT | R:TCTTGTACTCCGTGGCATCCT |
| 基因 Gene | 标签 Label | 链 Strand | 阅读框 Reading frame | 起始 Start | 终止 Stop | 长度 Length/bp |
|---|---|---|---|---|---|---|
| PsMATE14 | ORF2 | + | 1 | 361 | 1 785 | 1 425 |
| PsMATE16A | ORF7 | + | 3 | 66 | 1 535 | 1 470 |
| PsMATE16B | ORF8 | + | 3 | 90 | 1 589 | 1 500 |
| PsMATE23 | ORF1 | + | 1 | 469 | 1 956 | 1 488 |
| PsMATE27 | ORF9 | + | 3 | 276 | 1 754 | 1 479 |
| PsMATE32 | ORF6 | + | 3 | 39 | 1 653 | 1 614 |
| PsMATE33 | ORF4 | + | 2 | 167 | 1 612 | 1 446 |
| PsMATE35A | ORF1 | + | 1 | 40 | 1 521 | 1 482 |
| PsMATE35B | ORF6 | + | 3 | 150 | 1 622 | 1 473 |
| PsMATE40 | ORF9 | + | 3 | 234 | 1 790 | 1 557 |
| PsMATE41A | ORF3 | + | 2 | 32 | 1 609 | 1 578 |
| PsMATE41B | ORF5 | + | 3 | 51 | 1 577 | 1 527 |
| PsMATE41C | ORF3 | + | 1 | 98 | 1 609 | 1 512 |
| PsMATE48 | ORF2 | + | 2 | 95 | 1 744 | 1 650 |
| PsMATE49 | ORF1 | + | 1 | 34 | 1 539 | 1 506 |
| PsMATE51 | ORF1 | + | 2 | 194 | 1 831 | 1 638 |
| PsMATE56 | ORF3 | + | 2 | 230 | 1 753 | 1 524 |
Table 2 Analysis of PsMATE family gene sequence in the seeds of P. sibirica
| 基因 Gene | 标签 Label | 链 Strand | 阅读框 Reading frame | 起始 Start | 终止 Stop | 长度 Length/bp |
|---|---|---|---|---|---|---|
| PsMATE14 | ORF2 | + | 1 | 361 | 1 785 | 1 425 |
| PsMATE16A | ORF7 | + | 3 | 66 | 1 535 | 1 470 |
| PsMATE16B | ORF8 | + | 3 | 90 | 1 589 | 1 500 |
| PsMATE23 | ORF1 | + | 1 | 469 | 1 956 | 1 488 |
| PsMATE27 | ORF9 | + | 3 | 276 | 1 754 | 1 479 |
| PsMATE32 | ORF6 | + | 3 | 39 | 1 653 | 1 614 |
| PsMATE33 | ORF4 | + | 2 | 167 | 1 612 | 1 446 |
| PsMATE35A | ORF1 | + | 1 | 40 | 1 521 | 1 482 |
| PsMATE35B | ORF6 | + | 3 | 150 | 1 622 | 1 473 |
| PsMATE40 | ORF9 | + | 3 | 234 | 1 790 | 1 557 |
| PsMATE41A | ORF3 | + | 2 | 32 | 1 609 | 1 578 |
| PsMATE41B | ORF5 | + | 3 | 51 | 1 577 | 1 527 |
| PsMATE41C | ORF3 | + | 1 | 98 | 1 609 | 1 512 |
| PsMATE48 | ORF2 | + | 2 | 95 | 1 744 | 1 650 |
| PsMATE49 | ORF1 | + | 1 | 34 | 1 539 | 1 506 |
| PsMATE51 | ORF1 | + | 2 | 194 | 1 831 | 1 638 |
| PsMATE56 | ORF3 | + | 2 | 230 | 1 753 | 1 524 |
| 基因 Gene | 氨基酸数 Number of amino acid/AA | 相对分子质量 Molecular weight/Da | 理论等电点 pI | 不稳定指数 Instability index | 脂肪族指数 Aliphatic index | 总平均亲水系数 Grand average of hydropathicity | 磷酸化位点数 Numbers of phosphorylation sites |
|---|---|---|---|---|---|---|---|
| PsMATE14 | 474 | 51 553.06 | 6.86 | 31.31(Ⅱ) | 122.62 | 0.769 | 34 |
| PsMATE16A | 489 | 52 624.08 | 8.61 | 25.58(Ⅱ) | 118.45 | 0.632 | 30 |
| PsMATE16B | 499 | 54 117.13 | 7.94 | 27.96(Ⅱ) | 120.92 | 0.700 | 42 |
| PsMATE23 | 495 | 54 556.32 | 5.92 | 33.91(Ⅱ) | 122.63 | 0.662 | 38 |
| PsMATE27 | 492 | 54 308.24 | 6.21 | 29.20(Ⅱ) | 115.16 | 0.723 | 37 |
| PsMATE32 | 538 | 59 052.63 | 6.99 | 25.68(Ⅱ) | 114.20 | 0.640 | 31 |
| PsMATE33 | 481 | 51 857.24 | 5.13 | 27.00(Ⅱ) | 123.43 | 0.820 | 24 |
| PsMATE35A | 493 | 54 190.58 | 5.59 | 29.57(Ⅱ) | 126.61 | 0.891 | 32 |
| PsMATE35B | 490 | 53 825.66 | 5.38 | 31.25(Ⅱ) | 122.98 | 0.761 | 37 |
| PsMATE40 | 518 | 56 515.56 | 7.09 | 30.34(Ⅱ) | 120.29 | 0.676 | 40 |
| PsMATE41A | 525 | 57 331.33 | 6.01 | 28.65(Ⅱ) | 117.56 | 0.663 | 41 |
| PsMATE41B | 508 | 54 875.76 | 6.45 | 22.83(Ⅱ) | 123.25 | 0.722 | 35 |
| PsMATE41C | 503 | 54 680.77 | 6.52 | 23.51(Ⅱ) | 129.30 | 0.835 | 37 |
| PsMATE48 | 549 | 59 697.29 | 7.01 | 37.15(Ⅱ) | 110.87 | 0.485 | 48 |
| PsMATE49 | 501 | 54 511.57 | 8.11 | 37.21(Ⅱ) | 122.67 | 0.626 | 32 |
| PsMATE51 | 545 | 58 758.03 | 8.50 | 38.09(Ⅱ) | 113.32 | 0.517 | 56 |
| PsMATE56 | 507 | 54 725.26 | 8.26 | 32.54(Ⅱ) | 123.49 | 0.708 | 33 |
Table 3 Physical and chemical characters of PsMATE protein in the seeds of P. sibirica
| 基因 Gene | 氨基酸数 Number of amino acid/AA | 相对分子质量 Molecular weight/Da | 理论等电点 pI | 不稳定指数 Instability index | 脂肪族指数 Aliphatic index | 总平均亲水系数 Grand average of hydropathicity | 磷酸化位点数 Numbers of phosphorylation sites |
|---|---|---|---|---|---|---|---|
| PsMATE14 | 474 | 51 553.06 | 6.86 | 31.31(Ⅱ) | 122.62 | 0.769 | 34 |
| PsMATE16A | 489 | 52 624.08 | 8.61 | 25.58(Ⅱ) | 118.45 | 0.632 | 30 |
| PsMATE16B | 499 | 54 117.13 | 7.94 | 27.96(Ⅱ) | 120.92 | 0.700 | 42 |
| PsMATE23 | 495 | 54 556.32 | 5.92 | 33.91(Ⅱ) | 122.63 | 0.662 | 38 |
| PsMATE27 | 492 | 54 308.24 | 6.21 | 29.20(Ⅱ) | 115.16 | 0.723 | 37 |
| PsMATE32 | 538 | 59 052.63 | 6.99 | 25.68(Ⅱ) | 114.20 | 0.640 | 31 |
| PsMATE33 | 481 | 51 857.24 | 5.13 | 27.00(Ⅱ) | 123.43 | 0.820 | 24 |
| PsMATE35A | 493 | 54 190.58 | 5.59 | 29.57(Ⅱ) | 126.61 | 0.891 | 32 |
| PsMATE35B | 490 | 53 825.66 | 5.38 | 31.25(Ⅱ) | 122.98 | 0.761 | 37 |
| PsMATE40 | 518 | 56 515.56 | 7.09 | 30.34(Ⅱ) | 120.29 | 0.676 | 40 |
| PsMATE41A | 525 | 57 331.33 | 6.01 | 28.65(Ⅱ) | 117.56 | 0.663 | 41 |
| PsMATE41B | 508 | 54 875.76 | 6.45 | 22.83(Ⅱ) | 123.25 | 0.722 | 35 |
| PsMATE41C | 503 | 54 680.77 | 6.52 | 23.51(Ⅱ) | 129.30 | 0.835 | 37 |
| PsMATE48 | 549 | 59 697.29 | 7.01 | 37.15(Ⅱ) | 110.87 | 0.485 | 48 |
| PsMATE49 | 501 | 54 511.57 | 8.11 | 37.21(Ⅱ) | 122.67 | 0.626 | 32 |
| PsMATE51 | 545 | 58 758.03 | 8.50 | 38.09(Ⅱ) | 113.32 | 0.517 | 56 |
| PsMATE56 | 507 | 54 725.26 | 8.26 | 32.54(Ⅱ) | 123.49 | 0.708 | 33 |
| 蛋白Protein | α-螺旋α-helix | β-折叠β-sheet | 延伸链Extending strand | 无规卷曲Random coil |
|---|---|---|---|---|
| PsMATE14 | 320(67.51%) | 17(3.59%) | 57(12.03%) | 80(16.88%) |
| PsMATE16A | 295(60.33%) | 21(4.29%) | 64(13.09%) | 109(22.29%) |
| PsMATE16B | 289(57.92%) | 23(4.61%) | 67(13.43%) | 120(24.05%) |
| PsMATE23 | 296(59.80%) | 18(3.64%) | 75(15.15%) | 106(21.41%) |
| PsMATE27 | 298(60.57%) | 16(3.25%) | 71(14.43%) | 107(21.75%) |
| PsMATE32 | 302(56.13%) | 26(4.83%) | 78(14.50%) | 132(24.54%) |
| PsMATE33 | 294(61.12%) | 18(3.74%) | 76(15.80%) | 93(19.33%) |
| PsMATE35A | 308(62.47%) | 19(3.85%) | 63(12.78%) | 103(20.89%) |
| PsMATE35B | 274(55.92%) | 19(3.88%) | 88(17.96%) | 109(22.24%) |
| PsMATE40 | 310(59.85%) | 20(3.86%) | 72(13.90%) | 116(22.39%) |
| PsMATE41A | 340(64.76%) | 15(2.86%) | 66(12.57%) | 104(19.81%) |
| PsMATE41B | 305(60.04%) | 16(3.15%) | 74(14.57%) | 113(22.24%) |
| PsMATE41C | 306(60.83%) | 16(3.18%) | 80(15.90%) | 101(20.08%) |
| PsMATE48 | 307(55.92%) | 18(3.28%) | 65(11.84%) | 159(28.96%) |
| PsMATE49 | 298(59.48%) | 18(3.59%) | 59(11.78%) | 126(25.15%) |
| PsMATE51 | 308(56.51%) | 22(4.04%) | 68(12.48%) | 147(26.97%) |
| PsMATE56 | 310(61.14%) | 18(3.55%) | 54(10.65%) | 125(24.65%) |
Table 4 Secondary structure analysis of PsMATE protein in the seeds of P. sibirica
| 蛋白Protein | α-螺旋α-helix | β-折叠β-sheet | 延伸链Extending strand | 无规卷曲Random coil |
|---|---|---|---|---|
| PsMATE14 | 320(67.51%) | 17(3.59%) | 57(12.03%) | 80(16.88%) |
| PsMATE16A | 295(60.33%) | 21(4.29%) | 64(13.09%) | 109(22.29%) |
| PsMATE16B | 289(57.92%) | 23(4.61%) | 67(13.43%) | 120(24.05%) |
| PsMATE23 | 296(59.80%) | 18(3.64%) | 75(15.15%) | 106(21.41%) |
| PsMATE27 | 298(60.57%) | 16(3.25%) | 71(14.43%) | 107(21.75%) |
| PsMATE32 | 302(56.13%) | 26(4.83%) | 78(14.50%) | 132(24.54%) |
| PsMATE33 | 294(61.12%) | 18(3.74%) | 76(15.80%) | 93(19.33%) |
| PsMATE35A | 308(62.47%) | 19(3.85%) | 63(12.78%) | 103(20.89%) |
| PsMATE35B | 274(55.92%) | 19(3.88%) | 88(17.96%) | 109(22.24%) |
| PsMATE40 | 310(59.85%) | 20(3.86%) | 72(13.90%) | 116(22.39%) |
| PsMATE41A | 340(64.76%) | 15(2.86%) | 66(12.57%) | 104(19.81%) |
| PsMATE41B | 305(60.04%) | 16(3.15%) | 74(14.57%) | 113(22.24%) |
| PsMATE41C | 306(60.83%) | 16(3.18%) | 80(15.90%) | 101(20.08%) |
| PsMATE48 | 307(55.92%) | 18(3.28%) | 65(11.84%) | 159(28.96%) |
| PsMATE49 | 298(59.48%) | 18(3.59%) | 59(11.78%) | 126(25.15%) |
| PsMATE51 | 308(56.51%) | 22(4.04%) | 68(12.48%) | 147(26.97%) |
| PsMATE56 | 310(61.14%) | 18(3.55%) | 54(10.65%) | 125(24.65%) |
| 蛋白 Protein | 跨膜区 Transmembrane domain | 质膜 Plasma membrane | 液泡 Vacuolar | 高尔基体 Golgi | 内质网 Endoplasmic reticulum | 叶绿体 Chloroplast | 细胞质 Cytoplasmic | 细胞核 Nucleus |
|---|---|---|---|---|---|---|---|---|
| PsMATE14 | 10 | 6 | 7 | - | - | - | - | - |
| PsMATE16A | 12 | 6 | 5 | 1 | 2 | - | - | - |
| PsMATE16B | 12 | 6 | 5 | 1 | 2 | - | - | - |
| PsMATE23 | 12 | 7 | 3 | 3 | 1 | - | - | - |
| PsMATE27 | 10 | 12 | 2 | - | - | - | - | - |
| PsMATE32 | 12 | 8 | 3 | 2 | 1 | - | - | - |
| PsMATE33 | 12 | 13 | 1 | - | - | - | - | - |
| PsMATE35A | 12 | 9 | 4 | - | 1 | - | - | - |
| PsMATE35B | 11 | 5 | 4 | - | 3 | 1 | - | - |
| PsMATE40 | 12 | 7 | 4 | 2 | 1 | - | - | - |
| PsMATE41A | 12 | 4 | 8 | - | 2 | - | - | - |
| PsMATE41B | 12 | 10 | 1 | 2 | 1 | - | - | - |
| PsMATE41C | 9 | 8 | 4 | 1 | 1 | - | - | - |
| PsMATE48 | 11 | 10 | 2 | - | 2 | - | - | - |
| PsMATE49 | 11 | 11 | 2 | - | 1 | - | - | - |
| PsMATE51 | 10 | 6 | 3 | - | 3 | - | 1 | 1 |
| PsMATE56 | 10 | 8 | 2 | 3 | 1 | - | - | - |
Table 5 Analyses of transmembrane domains and subcellular localizations of PsMATE protein in the seeds of P. sibirica
| 蛋白 Protein | 跨膜区 Transmembrane domain | 质膜 Plasma membrane | 液泡 Vacuolar | 高尔基体 Golgi | 内质网 Endoplasmic reticulum | 叶绿体 Chloroplast | 细胞质 Cytoplasmic | 细胞核 Nucleus |
|---|---|---|---|---|---|---|---|---|
| PsMATE14 | 10 | 6 | 7 | - | - | - | - | - |
| PsMATE16A | 12 | 6 | 5 | 1 | 2 | - | - | - |
| PsMATE16B | 12 | 6 | 5 | 1 | 2 | - | - | - |
| PsMATE23 | 12 | 7 | 3 | 3 | 1 | - | - | - |
| PsMATE27 | 10 | 12 | 2 | - | - | - | - | - |
| PsMATE32 | 12 | 8 | 3 | 2 | 1 | - | - | - |
| PsMATE33 | 12 | 13 | 1 | - | - | - | - | - |
| PsMATE35A | 12 | 9 | 4 | - | 1 | - | - | - |
| PsMATE35B | 11 | 5 | 4 | - | 3 | 1 | - | - |
| PsMATE40 | 12 | 7 | 4 | 2 | 1 | - | - | - |
| PsMATE41A | 12 | 4 | 8 | - | 2 | - | - | - |
| PsMATE41B | 12 | 10 | 1 | 2 | 1 | - | - | - |
| PsMATE41C | 9 | 8 | 4 | 1 | 1 | - | - | - |
| PsMATE48 | 11 | 10 | 2 | - | 2 | - | - | - |
| PsMATE49 | 11 | 11 | 2 | - | 1 | - | - | - |
| PsMATE51 | 10 | 6 | 3 | - | 3 | - | 1 | 1 |
| PsMATE56 | 10 | 8 | 2 | 3 | 1 | - | - | - |
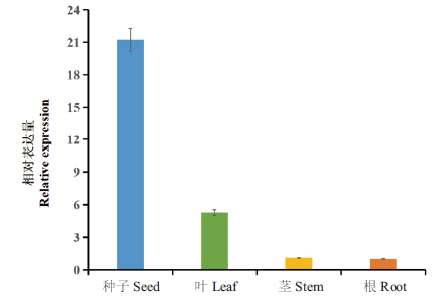
Fig. 8 Relative expressions of PsMATE40 gene in different parts of P. sibirica The lowest expression of PsMATE40 gene in the roots was taken as control,and standardized calibration is 1
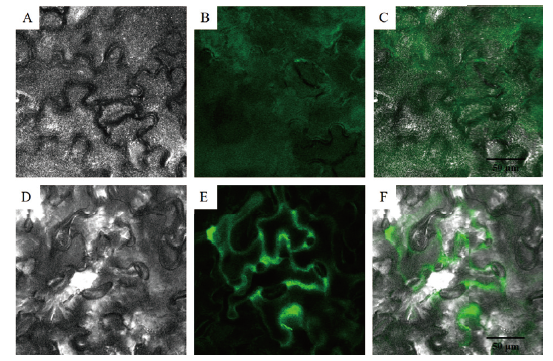
Fig. 9 Subcellular localization of PsMATE40 protein in the seeds of P. sibirica A is bright field image of the control. B is fluorescence image of the control. C is merged image of the control. D is bright field image of GFP-PsMATE40. E is fluorescence image of GFP-PsMATE40. F is merged image of GFP-PsMATE40
| [1] | 董胜君. 山杏种质资源遗传多样性及优特种质发掘研究[D]. 沈阳:沈阳农业大学, 2020. |
| Dong SJ. Genetic diversity of wild apricot germplasm resources and exploration of superior germplasm[D]. Shenyang:Shenyang Agricultural University, 2020. | |
| [2] | 焦中高, 吕真真, 刘杰超, 等. 苦杏仁去皮热风干燥适宜温度提高油脂品质[J]. 农业工程学报, 2016, 32(4): 262-268. |
| Jiao ZG, Lü ZZ, Liu JC, et al. Suitable hot air drying temperature improving quality of apricot kernel oil[J]. Trans Chin Soc Agric Eng, 2016, 32(4): 262-268. | |
| [3] |
Darbani B, Motawia MS, Olsen CE, et al. The biosynthetic gene cluster for the cyanogenic glucoside dhurrin in Sorghum bicolor contains its co-expressed vacuolar MATE transporter[J]. Sci Rep, 2016, 6: 37079.
doi: 10.1038/srep37079 URL |
| [4] |
Brown MH, Paulsen IT, Skurray RA. The multidrug efflux protein NorM is a prototype of a new family of transporters[J]. Mol Microbiol, 1999, 31(1): 394-395.
pmid: 9987140 |
| [5] |
Wang LH, Bei XJ, Gao JS, et al. The similar and different evolutionary trends of MATE family occurred between rice and Arabidopsis thaliana[J]. BMC Plant Biol, 2016, 16(1): 207.
doi: 10.1186/s12870-016-0895-0 URL |
| [6] |
Morita Y, Kodama K, Shiota S, et al. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli[J]. Antimicrob Agents Chemother, 1998, 42(7): 1778-1782.
pmid: 9661020 |
| [7] |
Diener AC, Gaxiola RA, Fink GR. Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins[J]. Plant Cell, 2001, 13(7): 1625-1638.
pmid: 11449055 |
| [8] |
Li L, He Z, Pandey GK, et al. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification[J]. J Biol Chem, 2002, 277(7): 5360-5368.
doi: 10.1074/jbc.M108777200 URL |
| [9] |
Li NN, Meng HJ, Xing HT, et al. Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus[J]. J Exp Bot, 2017, 68(20): 5669-5683.
doi: 10.1093/jxb/erx370 URL |
| [10] |
Li Y, He H, He LF. Genome-wide analysis of the MATE gene family in potato[J]. Mol Biol Rep, 2019, 46(1): 403-414.
doi: 10.1007/s11033-018-4487-y URL |
| [11] |
Dong BY, Niu LL, Meng D, et al. Genome-wide analysis of MATE transporters and response to metal stress in Cajanus cajan[J]. J Plant Interact, 2019, 14(1): 265-275.
doi: 10.1080/17429145.2019.1620884 URL |
| [12] |
Upadhyay N, Kar D, Deepak Mahajan B, et al. The multitasking abilities of MATE transporters in plants[J]. J Exp Bot, 2019, 70(18): 4643-4656.
doi: 10.1093/jxb/erz246 pmid: 31106838 |
| [13] |
Wang J, Lin W, Yin Z, et al. Comprehensive evaluation of fuel properties and complex regulation of intracellular transporters for high oil production in developing seeds of Prunus sibirica for woody biodiesel[J]. Biotechnol Biofuels, 2019, 12: 6.
doi: 10.1186/s13068-018-1347-x pmid: 30622648 |
| [14] |
Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites[J]. J Mol Biol, 1999, 294(5): 1351-1362.
pmid: 10600390 |
| [15] |
Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis[J]. Nat Protoc, 2015, 10(6): 845-858.
doi: 10.1038/nprot.2015.053 URL |
| [16] | Nielsen H. Predicting secretory proteins with SignalP[J]. Methods Mol Biol Clifton N J, 2017, 1611: 59-73. |
| [17] |
Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria[J]. Proteins, 1991, 11(2): 95-110.
pmid: 1946347 |
| [18] |
Niu J, Zhu B, Cai J, et al. Selection of reference genes for gene expression studies in Siberian Apricot(Prunus sibirica L.)Germplasm using quantitative real-time PCR[J]. PLoS One, 2014, 9(8): e103900.
doi: 10.1371/journal.pone.0103900 URL |
| [19] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method[J]. Methods, 2001, 25(4): 402-408.
pmid: 11846609 |
| [20] |
Niu J, An JY, Wang LB, et al. Transcriptomic analysis revealed the mechanism of oil dynamic accumulation during developing Siberian apricot(Prunus sibirica L.)seed kernels for the development of woody biodiesel[J]. Biotechnol Biofuels, 2015, 8: 29.
doi: 10.1186/s13068-015-0213-3 URL |
| [21] | 刘力力, 林子欣, 胡锦赫, 等. 山杏CAT家族基因的生物信息学预测及表达分析[J]. 分子植物育种, 2018, 16(22): 7255-7263. |
| Liu LL, Lin ZX, Hu JH, et al. Bioinformatic prediction and expression analysis of CAT gene family from Prunus sibirica L[J]. Mol Plant Breed, 2018, 16(22): 7255-7263. | |
| [22] |
Santos ALD, Chaves-Silva S, Yang L, et al. Global analysis of the MATE gene family of metabolite transporters in tomato[J]. BMC Plant Biol, 2017, 17(1): 185.
doi: 10.1186/s12870-017-1115-2 URL |
| [23] |
Min X, Jin X, Liu W, et al. Transcriptome-wide characterization and functional analysis of MATE transporters in response to aluminum toxicity in Medicago sativa L[J]. PeerJ, 2019, 7: e6302.
doi: 10.7717/peerj.6302 URL |
| [24] | 王作敏. 棕色棉纤维MATE基因的克隆及功能初步研究[D]. 石河子:石河子大学, 2018. |
| Wang ZM. Cloning and functional preliminary analysis of fiber MATE gene in brown cotton[D]. Shihezi:Shihezi University, 2018. | |
| [25] |
Kusakizako T, Miyauchi H, Ishitani R, et al. Structural biology of the multidrug and toxic compound extrusion superfamily transporters[J]. Biochim Biophys Acta Biomembr, 2020, 1862(12): 183154.
doi: 10.1016/j.bbamem.2019.183154 URL |
| [26] |
Chung YJ, Krueger C, Metzgar D, et al. Size comparisons among integral membrane transport protein homologues in bacteria, Archaea, and Eucarya[J]. J Bacteriol, 2001, 183(3): 1012-1021.
pmid: 11208800 |
| [27] |
Miyauchi H, Moriyama S, Kusakizako T, et al. Structural basis for xenobiotic extrusion by eukaryotic MATE transporter[J]. Nat Commun, 2017, 8(1): 1633.
doi: 10.1038/s41467-017-01541-0 URL |
| [28] |
Zhang HW, Zhao FG, Tang RJ, et al. Two tonoplast MATE proteins function as turgor-regulating chloride channels in Arabidopsis[J]. PNAS, 2017, 114(10): E2036-E2045.
doi: 10.1073/pnas.1616203114 URL |
| [29] |
Thompson EP, Wilkins C, Demidchik V, et al. An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development[J]. J Exp Bot, 2010, 61(2): 439-451.
doi: 10.1093/jxb/erp312 pmid: 19995827 |
| [30] |
Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, et al. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium[J]. Plant Cell, 2001, 13(4): 853-871.
pmid: 11283341 |
| [31] |
Marinova K, Pourcel L, Weder B, et al. The Arabidopsis MATE transporter TT12 Acts as a vacuolar flavonoid/H+ -antiporter active in proanthocyanidin-accumulating cells of the seed coat[J]. Plant Cell, 2007, 19(6): 2023-2038.
doi: 10.1105/tpc.106.046029 URL |
| [32] |
Seo PJ, Park J, Park MJ, et al. A Golgi-localized MATE transporter mediates iron homoeostasis under osmotic stress in Arabidopsis[J]. Biochem J, 2012, 442(3): 551-561.
doi: 10.1042/BJ20111311 URL |
| [33] |
Burko Y, Geva Y, Refael-Cohen A, et al. From organelle to organ:ZRIZI MATE-Type transporter is an organelle transporter that enhances organ initiation[J]. Plant Cell Physiol, 2011, 52(3): 518-527.
doi: 10.1093/pcp/pcr007 URL |
| [34] |
Wang R, Liu X, Liang S, et al. A subgroup of MATE transporter genes regulates hypocotyl cell elongation in Arabidopsis[J]. J Exp Bot, 2015, 66(20): 6327-6343.
doi: 10.1093/jxb/erv344 pmid: 26160579 |
| [35] |
Li R, Li J, Li S, et al. ADP1 affects plant architecture by regulating local auxin biosynjournal[J]. PLoS Genet, 2014, 10(1): e1003954.
doi: 10.1371/journal.pgen.1003954 URL |
| [36] |
Tian W, Hou C, Ren Z, et al. A molecular pathway for CO2 response in Arabidopsis guard cells[J]. Nat Commun, 2015, 6: 6057.
doi: 10.1038/ncomms7057 URL |
| [37] |
Rogers EE, Guerinot ML. FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis[J]. Plant Cell, 2002, 14(8): 1787-1799.
doi: 10.1105/tpc.001495 URL |
| [38] | Kovinich N, Wang YQ, Adegboye J, et al. Arabidopsis MATE 45 antagonizes local abscisic acid signaling to mediate development and abiotic stress responses[J]. Plant Direct, 2018, 2(10): e00087. |
| [39] |
Yamasaki K, Motomura Y, Yagi Y, et al. Chloroplast envelope localization of EDS5, an essential factor for salicylic acid biosynjournal in Arabidopsis thaliana[J]. Plant Signal Behav, 2013, 8(4): e23603.
doi: 10.4161/psb.23603 URL |
| [40] |
Serrano M, Wang B, Aryal B, et al. Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5[J]. Plant Physiol, 2013, 162(4): 1815-1821.
doi: 10.1104/pp.113.218156 pmid: 23757404 |
| [41] |
Rekhter D, Lüdke D, Ding Y, et al. Isochorismate-derived biosynjournal of the plant stress hormone salicylic acid[J]. Science, 2019, 365(6452): 498-502.
doi: 10.1126/science.aaw1720 URL |
| [42] | Ishihara T, Sekine KT, Hase S, et al. Overexpression of the Arabidopsis thaliana EDS5 gene enhances resistance to viruses[J]. Plant Biol:Stuttg, 2008, 10(4): 451-461. |
| [43] |
Liu J, Li Y, Wang W, et al. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean[J]. BMC Genomics, 2016, 17: 223.
doi: 10.1186/s12864-016-2559-8 URL |
| [44] | Ali E, Saand MA, Khan AR, et al. Genome-wide identification and expression analysis of detoxification efflux carriers(DTX)genes family under abiotic stresses in flax[J]. Physiol Plant, 2021, 171(4): 483-501. |
| [45] |
Santamour FS Jr. Amygdalin in Prunus leaves[J]. Phytochemistry, 1998, 47(8): 1537-1538.
doi: 10.1016/S0031-9422(97)00787-5 URL |
| [46] |
London-Shafir I, Shafir S, Eisikowitch D. Amygdalin in almond nectar and pollen - facts and possible roles[J]. Plant Syst Evol, 2003, 238(1/2/3/4): 87-95.
doi: 10.1007/s00606-003-0272-y URL |
| [47] |
Tiwari M, Sharma D, Singh M, et al. Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis[J]. Sci Rep, 2014, 4: 3964.
doi: 10.1038/srep03964 URL |
| [1] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [2] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [3] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [4] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [5] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [6] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [7] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [8] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [9] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [10] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [11] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [12] | GE Wen-dong, WANG Teng-hui, MA Tian-yi, FAN Zhen-yu, WANG Yu-shu. Genome-wide Identification of the PRX Gene Family in Cabbage(Brassica oleracea L. var. capitata)and Expression Analysis Under Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 252-260. |
| [13] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [14] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [15] | YOU Chui-huai, XIE Jin-jin, ZHANG Ting, CUI Tian-zhen, SUN Xin-lu, ZANG Shou-jian, WU Yi-ning, SUN Meng-yao, QUE You-xiong, SU Ya-chun. Identification of the Lipoxygenase Gene GeLOX1 and Expression Analysis Under Low Temperature Stress in Gelsmium elegans [J]. Biotechnology Bulletin, 2023, 39(11): 318-327. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||