








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (11): 212-224.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0108
Previous Articles Next Articles
ZHANG Yun-chuan( ), LIN Yi-xuan, CAO Xin-wen, WANG Hai-nan, YAN Jie(
), LIN Yi-xuan, CAO Xin-wen, WANG Hai-nan, YAN Jie( )
)
Received:2021-01-27
Online:2021-11-26
Published:2021-12-03
Contact:
YAN Jie
E-mail:1761241940@qq.com;1653842328@qq.com
ZHANG Yun-chuan, LIN Yi-xuan, CAO Xin-wen, WANG Hai-nan, YAN Jie. TkDREB2 Clone from Taraxacum kok-saghyz and Drought Tolerance Analysis of Transgenic Nicotiana tabacum[J]. Biotechnology Bulletin, 2021, 37(11): 212-224.

Fig. 1 Heat map of differentially expressed AP2/EREBP(A),bHLH(B),MYB(C)and WRKY(D)transcription factors Red expression is up-regulated,blue expression is down-regulated,the darker the color,the higher the expression level. MJ0_R,MJ6_R and MJ24_R indicate jasmonic acid treatment 0,6 and 24 h
| 基因信息Genetic information | 茉莉酸处理时间Treatment time by jasmonic acid | 差异表达倍数Log2FoldChange | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 基因名称 Gene name | 基因Gene ID | MJ6 | MJ0 | MJ24 | 6 VS 0 | P value | 24 VS 0 | P value | |||
| OPR | c49640_g1 | 25 210.72 | 956.65 | 4.72 | 1.71E-35 | ||||||
| 1 017.64 | 16 350.86 | 4.01 | 1.77E-31 | ||||||||
| c49640_g2 | 15 311.67 | 589.34 | 4.70 | 3.61E-35 | |||||||
| 626.95 | 10 019.59 | 4.00 | 3.15E-33 | ||||||||
| c52666_g1 | 2 642.99 | 268.00 | 3.30 | 1.82E-19 | |||||||
| 284.96 | 2 252.14 | 2.98 | 4.39E-20 | ||||||||
| AOS | c39329_g1 | 1 825.74 | 17.73 | 6.69 | 2.28E-17 | ||||||
| 18.87 | 10 973.08 | 9.18 | 3.60E-25 | ||||||||
| c47723_g1 | 603.38 | 7 806.60 | 3.69 | 1.46E-06 | |||||||
| LOX | c53508_g1 | 21 905.05 | 359.65 | 5.93 | 4.42E-10 | ||||||
| 382.62 | 49 808.79 | 7.02 | 2.75E-13 | ||||||||
| c51038_g1 | 294.97 | 895.27 | 1.60 | 1.62E-05 | |||||||
| AOC | c44998_g1 | 369.99 | 146.00 | 1.34 | 0.000 269 | ||||||
| 155.26 | 737.86 | 2.25 | 9.16E-12 | ||||||||
Table 1 Differential expression of genes encoding enzymes related to jasmonic acid synthesis
| 基因信息Genetic information | 茉莉酸处理时间Treatment time by jasmonic acid | 差异表达倍数Log2FoldChange | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 基因名称 Gene name | 基因Gene ID | MJ6 | MJ0 | MJ24 | 6 VS 0 | P value | 24 VS 0 | P value | |||
| OPR | c49640_g1 | 25 210.72 | 956.65 | 4.72 | 1.71E-35 | ||||||
| 1 017.64 | 16 350.86 | 4.01 | 1.77E-31 | ||||||||
| c49640_g2 | 15 311.67 | 589.34 | 4.70 | 3.61E-35 | |||||||
| 626.95 | 10 019.59 | 4.00 | 3.15E-33 | ||||||||
| c52666_g1 | 2 642.99 | 268.00 | 3.30 | 1.82E-19 | |||||||
| 284.96 | 2 252.14 | 2.98 | 4.39E-20 | ||||||||
| AOS | c39329_g1 | 1 825.74 | 17.73 | 6.69 | 2.28E-17 | ||||||
| 18.87 | 10 973.08 | 9.18 | 3.60E-25 | ||||||||
| c47723_g1 | 603.38 | 7 806.60 | 3.69 | 1.46E-06 | |||||||
| LOX | c53508_g1 | 21 905.05 | 359.65 | 5.93 | 4.42E-10 | ||||||
| 382.62 | 49 808.79 | 7.02 | 2.75E-13 | ||||||||
| c51038_g1 | 294.97 | 895.27 | 1.60 | 1.62E-05 | |||||||
| AOC | c44998_g1 | 369.99 | 146.00 | 1.34 | 0.000 269 | ||||||
| 155.26 | 737.86 | 2.25 | 9.16E-12 | ||||||||

Fig. 3 Recombinant plasmid pMD19T-TkDREB2 digestion M: DNA marker Ⅲ. 1-2: BamHI and Pst I double digestion pMD19T-TkDREB2 recombinant plasmid. 3: pMD19T-TkDREB2 recombinant plasmid
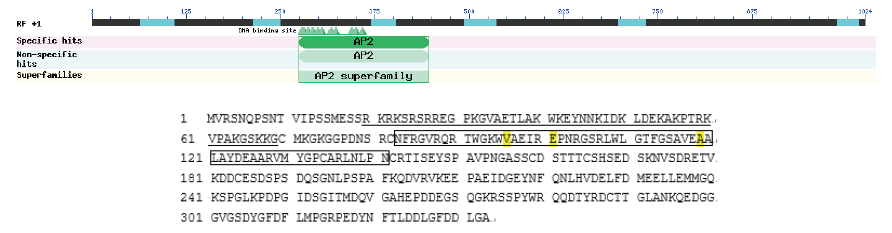
Fig. 8 TkDREB2 amino acid sequence analysis The sequence in the square represents the AP2 domain, and the underlined part represents the predicted nuclear localization signal sequence

Fig. 10 Phylogenetic tree and conservative motif analysis of TkDREB2 protein A:Phylogenetic tree and conservative motif analysis of TkDREB2 protein. B:5 conservative motif fragments. Taraxacum kok-saghyz:TkDREB2(Taraxacum kok-saghyz,ASL72071.1); Lactuca sativa:LsDREB2A(Lactuca sativa,XP_023748594.1); Cichorium intybus:CiDREB2(Cichorium intybus,AHJ08574.1); Cynara cardunculus:CcDREB2A(Cynara cardunculus,XP_024968974.1); Chrysanthemum vestitum:CvDREB2(Chrysanthemum vestitum,ABR23508.1); Helianthus annuus:HaDREB2A(Helianthus annuus,XP_021982098.1); Nicotiana sylvestris:NsDREB2A(Nicotiana sylvestris,XP_009788693.1); Nicotiana attenuata:NaDREB2A(Nicotiana attenuata,XP_019245341.1); Nicotiana tomentosiformis:NtDREB2A(Nicotiana tomentosiformis,XP_009594350.1); Nicotiana tabacum:NtDREB2A(Nicotiana tabacum,XP_016461559.1); Sesamum indicum:SiDREB2C(Sesamum indicum,XP_011084866.1)

Fig. 11 Recombinant plasmid pCAMBIA2300-35S-TkDREB2 digestion M: DNA marker Ⅲ; 1: pCAMBIA2300-35S-TkDREB2 recombinant plasmid;2-3: BamHI and Pst I double digestion pCAMBIA2300-35S-TkDREB2 recombinant plasmid

Fig. 13 Wild-type(WT)and genetically modified tobacco under normal growth conditions(A)and 22 d of drought treatment(B) Transgenic lines:Genetically modified tobacco;WT:wild type tobacco
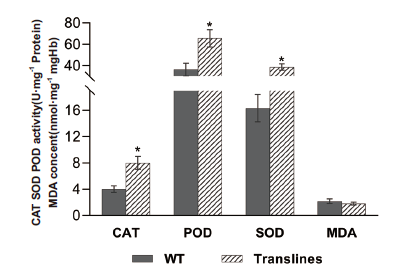
Fig. 14 SOD,POD and CAT enzyme activities and MDA content measurement results of wild-type and transgenic plants under normal growth conditions The mean measurement results of the physiological indexes of 3 wild-type and 3 transgenic tobaccos are compared by plotting. * indicates significant difference compared with wild-type(P<0.05,t-test),the same below

Fig. 16 Electrical conductivity measurement results of wild-type and transgenic plants after normal growth and drought treatment Transline 1-3:3 strains of genetically modified tobacco. The electrical conductivity of the wild-type plant is the mean value of the measurement results of 3 wild-type plants
| [1] |
Chini A, Fonseca S, Fernández G, et al. The JAZ family of repressors is the missing link in jasmonate signalling[J]. Nature, 2007, 448(7154): 666-671.
doi: 10.1038/nature06006 URL |
| [2] |
Fonseca S, Chini A, Hamberg M, et al. (+)-7-Iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate[J]. Nat Chem Biol, 2009, 5(5): 344-350.
doi: 10.1038/nchembio.161 pmid: 19349968 |
| [3] |
Westfall CS, Zubieta C, Herrmann J, et al. Structural basis for prereceptor modulation of plant hormones by GH3 proteins[J]. Science, 2012, 336(6089): 1708-1711.
doi: 10.1126/science.1221863 pmid: 22628555 |
| [4] |
Pauwels L, Barbero GF, Geerinck J, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling[J]. Nature, 2010, 464(7289): 788-791.
doi: 10.1038/nature08854 URL |
| [5] | 文锦芬, 赵凯, 邓明华. 转录因子DREB、ERF和NAC在介导植物响应生物和非生物胁迫中的作用[J]. 湖南生态科学学报, 2019, 6(3): 51-59. |
| Wen JF, Zhao K, Deng MH. Roles of transcription factors DREB, ERF and NAC in mediating plant responses to biotic and abiotic stresses[J]. J Hunan Ecol Sci, 2019, 6(3): 51-59. | |
| [6] | 王舟, 刘建秀. DREB/CBF类转录因子研究进展及其在草坪草和牧草抗逆基因工程中的应用[J]. 草业学报, 2011, 20(1): 222-236. |
| Wang Z, Liu JX. Advances in studies on DREB/CBF transcription factors, and their applications in genetic engineering for stress tolerance of turf and forage grasses[J]. Acta Prataculturae Sin, 2011, 20(1): 222-236. | |
| [7] |
Hussain SS, Kayani MA, Amjad M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants[J]. Biotechnol Prog, 2011, 27(2): 297-306.
doi: 10.1002/btpr.v27.2 URL |
| [8] |
Zhang S, Zhu C, Lyu Y, et al. Genome-wide identification, molecular evolution, and expression analysis provide new insights into the APETALA2/ethylene responsive factor(AP2/ERF)superfamily in Dimocarpus longan Lour[J]. BMC Genomics, 2020, 21(1): 62.
doi: 10.1186/s12864-020-6469-4 URL |
| [9] |
Aya K, Hobo T, et al. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway[J]. Plant Cell Physiol, 2014, 55(5): 897-912.
doi: 10.1093/pcp/pcu023 URL |
| [10] |
Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana[J]. Science, 2003, 301(5633): 653-657.
pmid: 12893945 |
| [11] |
Hu YX, Wang YH, Liu XF, et al. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development[J]. Cell Res, 2004, 14(1): 8-15.
doi: 10.1038/sj.cr.7290197 URL |
| [12] |
Sakuma Y, Liu Q, Dubouzet JG, et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression[J]. Biochem Biophys Res Commun, 2002, 290(3): 998-1009.
doi: 10.1006/bbrc.2001.6299 URL |
| [13] |
Nakano T, Suzuki K, Fujimura T, et al. Genome-wide analysis of the ERF gene family in Arabidopsis and rice[J]. Plant Physiol, 2006, 140(2): 411-432.
doi: 10.1104/pp.105.073783 URL |
| [14] |
Dossa K, Wei X, et al. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress[J]. BMC Plant Biol, 2016, 16(1): 171.
doi: 10.1186/s12870-016-0859-4 URL |
| [15] |
Ito Y, Katsura K, Maruyama K, et al. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice[J]. Plant Cell Physiol, 2006, 47(1): 141-153.
doi: 10.1093/pcp/pci230 URL |
| [16] |
Dubouzet JG, Sakuma Y, Ito Y, et al. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression[J]. Plant J, 2003, 33(4): 751-763.
pmid: 12609047 |
| [17] |
Qin F, Sakuma Y, Li J, et al. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L[J]. Plant Cell Physiol, 2004, 45(8): 1042-1052.
doi: 10.1093/pcp/pch118 URL |
| [18] |
Chen M, Xu Z, et al. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean(Glycine max L.)[J]. J Exp Bot, 2009, 60(1): 121-135.
doi: 10.1093/jxb/ern269 URL |
| [19] |
Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance[J]. J Exp Bot, 2007, 58(2): 221-227.
pmid: 17075077 |
| [20] |
Gilmour SJ, Sebolt AM, Salazar MP, et al. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation[J]. Plant Physiol, 2000, 124(4): 1854-1865.
pmid: 11115899 |
| [21] | Zong JM, Li XW, Zhou YH, et al. The AaDREB1 transcription factor from the cold-tolerant plant Adonis amurensis enhances abiotic stress tolerance in transgenic plant[J]. Int J Mol Sci, 2016, 17(4): E611. |
| [22] | Liu BJ, Zhou Y, Lan W, et al. LlDREB1G, a novel DREB subfamily gene from Lilium longiflorum, can enhance transgenic Arabidopsis tolerance to multiple abiotic stresses[J]. Plant Cell Tissue Organ Cult PCTOC, 2019, 138(3): 489-506. |
| [23] |
Knight H, Zarka DG, Okamoto H, et al. Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element[J]. Plant Physiol, 2004, 135(3): 1710-1717.
doi: 10.1104/pp.104.043562 URL |
| [24] |
Licausi F, Ohme-Takagi M, Perata P. APETALA2/Ethylene responsive factor(AP2/ERF)transcription factors:mediators of stress responses and developmental programs[J]. New Phytol, 2013, 199(3): 639-649.
doi: 10.1111/nph.12291 URL |
| [25] |
Liu Y, Zhao TJ, Liu JM, et al. The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box[J]. FEBS Lett, 2006, 580(5): 1303-1308.
doi: 10.1016/j.febslet.2006.01.048 URL |
| [26] | 高文俊, 徐静, 谢开云, 等. Na2CO3和NaHCO3胁迫下冰草的生长及生理响应[J]. 草业学报, 2011, 20(4): 299-304. |
| Gao WJ, Xu J, Xie KY, et al. Physiological responses of Agropyron cristatum under Na2CO3 and NaHCO3 stress[J]. Acta Prataculturae Sin, 2011, 20(4): 299-304. | |
| [27] |
Sakuma Y, Maruyama K, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression[J]. Plant Cell, 2006, 18(5): 1292-1309.
pmid: 16617101 |
| [28] |
Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance[J]. Trends Plant Sci, 2015, 20(4): 219-229.
doi: 10.1016/j.tplants.2015.02.001 URL |
| [29] |
Alam MM, Nahar K, Hasanuzzaman M, et al. Exogenous jasmonic acid modulates the physiology, antioxidant defense and glyoxalase systems in imparting drought stress tolerance in different Brassica species[J]. Plant Biotechnol Rep, 2014, 8(3): 279-293.
doi: 10.1007/s11816-014-0321-8 URL |
| [30] | Yosefi A, Mozafari AA, Javadi T. Jasmonic acid improved in vitro strawberry(Fragaria × ananassa Duch. )resistance to PEG-induced water stress[J]. Plant Cell Tissue Organ Cult PCTOC, 2020, 142(3): 549-558. |
| [31] |
Seo JS, Joo J, Kim MJ, et al. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice[J]. Plant J, 2011, 65(6): 907-921.
doi: 10.1111/tpj.2011.65.issue-6 URL |
| [32] |
Cao X, Yan J, Lei J, et al. De novo transcriptome sequencing of MeJA-induced Taraxacum koksaghyz Rodin to identify genes related to rubber formation[J]. Sci Rep, 2017, 7(1): 15697.
doi: 10.1038/s41598-017-14890-z URL |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [3] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [4] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [5] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [6] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [7] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [8] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [9] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [10] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [11] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [12] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [13] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [14] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [15] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||