








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (1): 77-85.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0181
Previous Articles Next Articles
LV Di( ), CHEN Ru-mei, ZHOU Xiao-jin(
), CHEN Ru-mei, ZHOU Xiao-jin( )
)
Received:2021-02-13
Online:2022-01-26
Published:2022-02-22
Contact:
ZHOU Xiao-jin
E-mail:lvdi1@foxmail.com;zhouxiaojin@caas.cn
LV Di, CHEN Ru-mei, ZHOU Xiao-jin. Interactions Between ZmJAZ and ZmMYC2 Using Bimolecular Fluorescence Complementation Assay[J]. Biotechnology Bulletin, 2022, 38(1): 77-85.
| 基因名称 Gene name | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| ZmActin | ZmActin-qF | ATGTTTCCTGGGATTGCCGAT |
| ZmActin-qR | CCAGTTTCGTCATACTCTCCCTTG | |
| ZmJAZ4 | ZmJAZ4-qF | CGCTCAGGCTCTGAAACAGTGAAACC |
| ZmJAZ4-qR | CGACGACAGACACAGTGCCTAAGAAT | |
| ZmJAZ8 | ZmJAZ8-qF | TTGAGAAACGCCGTGACAGGGT |
| ZmJAZ8-qR | GTCAGCCTTCACCGATGGGACT | |
| ZmJAZ12 | ZmJAZ12-qF | CTGATGCTAAGAAGCCTACTCGC |
| ZmJAZ12-qR | GCGTCTGAAGGAGAAGTTTGGTA |
Table 1 Primers used in qRT-PCR
| 基因名称 Gene name | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| ZmActin | ZmActin-qF | ATGTTTCCTGGGATTGCCGAT |
| ZmActin-qR | CCAGTTTCGTCATACTCTCCCTTG | |
| ZmJAZ4 | ZmJAZ4-qF | CGCTCAGGCTCTGAAACAGTGAAACC |
| ZmJAZ4-qR | CGACGACAGACACAGTGCCTAAGAAT | |
| ZmJAZ8 | ZmJAZ8-qF | TTGAGAAACGCCGTGACAGGGT |
| ZmJAZ8-qR | GTCAGCCTTCACCGATGGGACT | |
| ZmJAZ12 | ZmJAZ12-qF | CTGATGCTAAGAAGCCTACTCGC |
| ZmJAZ12-qR | GCGTCTGAAGGAGAAGTTTGGTA |
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 扩增产物 Product |
|---|---|---|
| ZmMYC2-F | ATGAACCTGTGGACGGAC | ZmMYC2-ORF |
| ZmMYC2-R | TTACCTGCCCATGGCAGACCCG | |
| ZmJAZ4-F | ATGGCCGCCTCCGGGAACAACA | ZmJAZ4-ORF |
| ZmJAZ4-R | TCAGAGCCTGAGCGCGAGCCACGA | |
| ZmJAZ8-F | ATGGATCTGTTGGAGCGGAATATT | ZmJAZ8-ORF |
| ZmJAZ8-R | TTACTGGGCCTTGCCCTCCGCTGT | |
| ZmJAZ12-F | ATGGCGGCGTCCGCGAGGCCCG | ZmJAZ12-ORF |
| ZmJAZ12-R | CTAGCTCAGGTTCAGGTTGGACTT | |
| ZmMYC2-GF | TCATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGACGACAA | ZmMYC2-GFP |
| ZmMYC2-GR | CCTCGCCCTTGCTCACCATTCTAGACCTGCCCATGGCAGACCCGGGCT | |
| ZmJAZ4-GF | ATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAACA | ZmJAZ4-GFP |
| ZmJAZ4-GR | CCTCGCCCTTGCTCACCATTCTAGAGAGCCTGAGCGCGAGCCAC | |
| ZmJAZ8-GF | ATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAATA | ZmJAZ8- GFP |
| ZmJAZ8-GR | CCTCGCCCTTGCTCACCATTCTAGACTGGGCCTTGCCCTCCGCT | |
| ZmJAZ12-GF | ATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-GFP |
| ZmJAZ12-GR | CCTCGCCCTTGCTCACCATCTAGAGCTCAGGTTCAGGTTGGACT | |
| ZmMYC2-NF | ATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGAC | ZmMYC2-YFPN |
| ZmMYC2-NR | CTTTTGCTCCATTCTAGACCTGCCCATGGCAGACCC | |
| ZmMYC2-NF | ATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGAC | ZmMYC2-YFPC |
| ZmMYC2-CR | GTATGGGTACATTCTAGACCTGCCCATGGCAGACCC | |
| ZmJAZ4-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAA | ZmJAZ4-YFPN |
| ZmJAZ4-NR | AACTTTTGCTCCATTCTAGAGAGCCTGAGCGCGAGCCACGAGG | |
| ZmJAZ4-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAA | ZmJAZ4-YFPC |
| ZmJAZ4-CR | CATCGTATGGGTACATTCTAGAGAGCCTGAGCGCGAGCCACGAGG | |
| ZmJAZ8-NF | CATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAAT | ZmJAZ8-YFPN |
| ZmJAZ8-NR | AACTTTTGCTCCATTCTAGACTGGGCCTTGCCCTCCGCTGTCAC | |
| ZmJAZ8-NF | CATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAAT | ZmJAZ8-YFPC |
| ZmJAZ8-CR | CATCGTATGGGTACATTCTAGACTGGGCCTTGCCCTCCGCTGTCAC | |
| ZmJAZ12-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-YFPN |
| ZmJAZ12-NR | AACTTTTGCTCCATTCTAGAGCTCAGGTTCAGGTTGGACTTGAC | |
| ZmJAZ12-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-YFPC |
| ZmJAZ12-CR | CATCGTATGGGTACATTCTAGAGCTCAGGTTCAGGTTGGACTTGAC |
Table 2 Primers used for gene cloning
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 扩增产物 Product |
|---|---|---|
| ZmMYC2-F | ATGAACCTGTGGACGGAC | ZmMYC2-ORF |
| ZmMYC2-R | TTACCTGCCCATGGCAGACCCG | |
| ZmJAZ4-F | ATGGCCGCCTCCGGGAACAACA | ZmJAZ4-ORF |
| ZmJAZ4-R | TCAGAGCCTGAGCGCGAGCCACGA | |
| ZmJAZ8-F | ATGGATCTGTTGGAGCGGAATATT | ZmJAZ8-ORF |
| ZmJAZ8-R | TTACTGGGCCTTGCCCTCCGCTGT | |
| ZmJAZ12-F | ATGGCGGCGTCCGCGAGGCCCG | ZmJAZ12-ORF |
| ZmJAZ12-R | CTAGCTCAGGTTCAGGTTGGACTT | |
| ZmMYC2-GF | TCATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGACGACAA | ZmMYC2-GFP |
| ZmMYC2-GR | CCTCGCCCTTGCTCACCATTCTAGACCTGCCCATGGCAGACCCGGGCT | |
| ZmJAZ4-GF | ATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAACA | ZmJAZ4-GFP |
| ZmJAZ4-GR | CCTCGCCCTTGCTCACCATTCTAGAGAGCCTGAGCGCGAGCCAC | |
| ZmJAZ8-GF | ATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAATA | ZmJAZ8- GFP |
| ZmJAZ8-GR | CCTCGCCCTTGCTCACCATTCTAGACTGGGCCTTGCCCTCCGCT | |
| ZmJAZ12-GF | ATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-GFP |
| ZmJAZ12-GR | CCTCGCCCTTGCTCACCATCTAGAGCTCAGGTTCAGGTTGGACT | |
| ZmMYC2-NF | ATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGAC | ZmMYC2-YFPN |
| ZmMYC2-NR | CTTTTGCTCCATTCTAGACCTGCCCATGGCAGACCC | |
| ZmMYC2-NF | ATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGAC | ZmMYC2-YFPC |
| ZmMYC2-CR | GTATGGGTACATTCTAGACCTGCCCATGGCAGACCC | |
| ZmJAZ4-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAA | ZmJAZ4-YFPN |
| ZmJAZ4-NR | AACTTTTGCTCCATTCTAGAGAGCCTGAGCGCGAGCCACGAGG | |
| ZmJAZ4-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAA | ZmJAZ4-YFPC |
| ZmJAZ4-CR | CATCGTATGGGTACATTCTAGAGAGCCTGAGCGCGAGCCACGAGG | |
| ZmJAZ8-NF | CATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAAT | ZmJAZ8-YFPN |
| ZmJAZ8-NR | AACTTTTGCTCCATTCTAGACTGGGCCTTGCCCTCCGCTGTCAC | |
| ZmJAZ8-NF | CATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAAT | ZmJAZ8-YFPC |
| ZmJAZ8-CR | CATCGTATGGGTACATTCTAGACTGGGCCTTGCCCTCCGCTGTCAC | |
| ZmJAZ12-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-YFPN |
| ZmJAZ12-NR | AACTTTTGCTCCATTCTAGAGCTCAGGTTCAGGTTGGACTTGAC | |
| ZmJAZ12-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-YFPC |
| ZmJAZ12-CR | CATCGTATGGGTACATTCTAGAGCTCAGGTTCAGGTTGGACTTGAC |
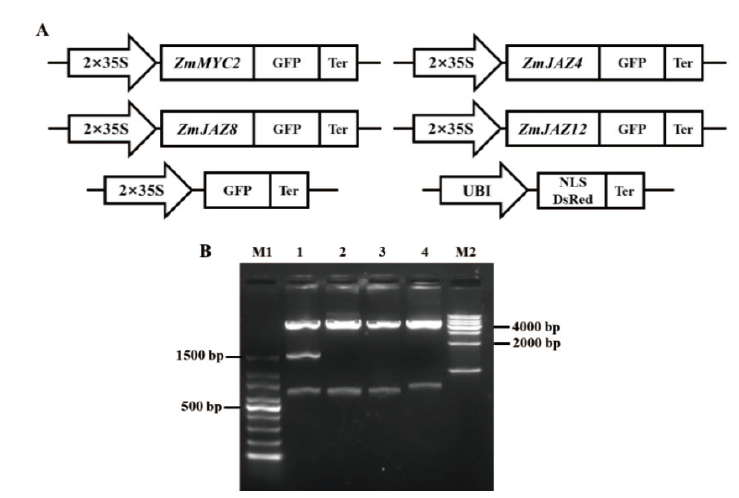
Fig.2 Vectors for subcellular localization and enzyme digestion identification A:Schematic diagram of vectors for subcellular localization,including control GFP vector and cell nucleus localization marker vector. B:Enzyme digestion identification of Xho I + Xba I in subcellular localization vectors. M1:100 bp DNA ladder. M2:1000 bp DNA ladder. Lane 1:pZmMYC2-GFP,lane 2:pZmJAZ4-GFP,lane 3:pZmJAZ8-GFP,and lane 4:pZmJAZ12-GFP
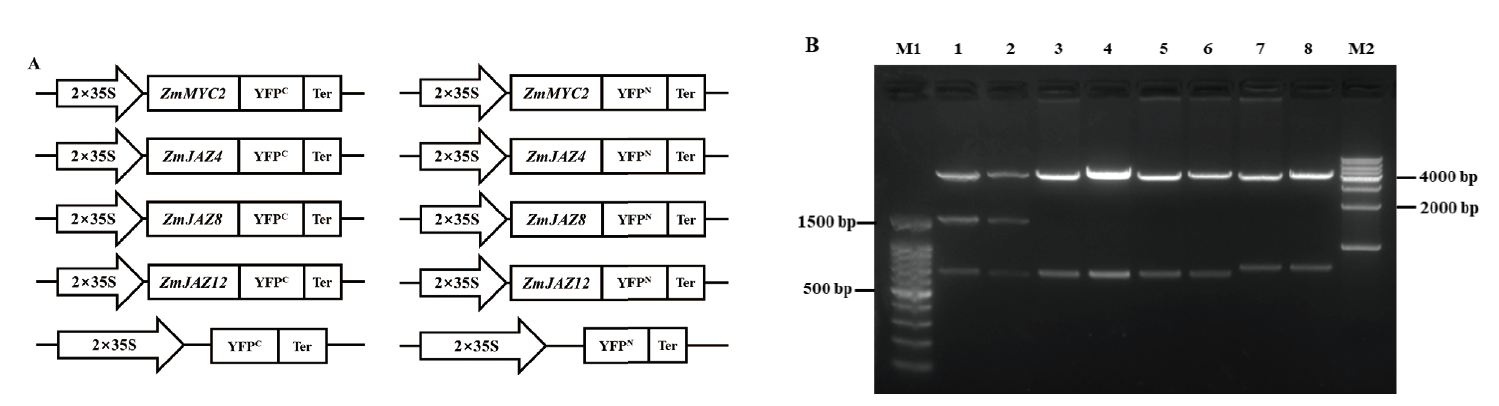
Fig.3 Vectors for BiFC assay and enzyme digestion identification A:Schematic diagram of vectors for BiFC assay. B:Enzyme digestion identification of BiFC vectors by Xho I + Xba I. M1:100 bp DNA ladder. M2:1 000 bp DNA ladder. Lane 1:pZmMYC2-YFPN,lane 2:pZmMYC2-YFPC,lane 3:pZmJAZ4-YFPN,lane 4:pZmJAZ4-YFPC,lane 5:pZmJAZ8-YFPN,lane 6:pZmJAZ8-YFPC,lane 7:pZmJAZ12-YFPN,and lane 8:pZmJAZ12-YFPC
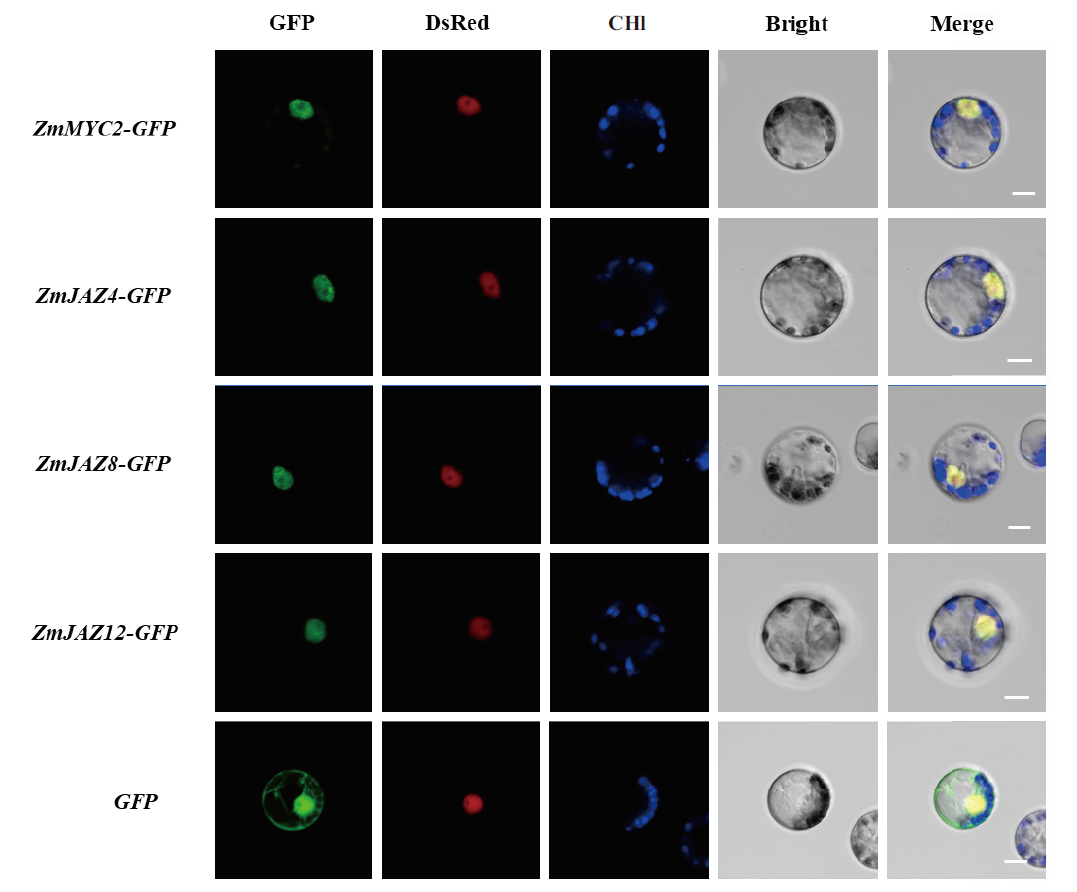
Fig. 4 Subcellular localization of ZmJAZ4,ZmJAZ8,ZmJAZ12 and ZmMYC2 The ORFs of ZmJAZ4、ZmJAZ8、ZmJAZ12 and ZmMYC2 were fused at the N terminal of GFP. GFP indicates the target protein fluorescence channel. DsRed indicates fluorescence channel for a nuclear localization maker. Chl indicates the chloroplast spontaneous fluorescence channel. Bright indicates the bright field. The Merge indicates channel combined the GFP,DsRed,Chl and the bright field. Scale bar = 10 μm

Fig. 5 BiFC assay to verify interactions between ZmJAZs with ZmMYC2 ZmMYC2 and 3 to-be-verified genes were constructed into the N-terminal and C-terminal of YFP for BiFC assay. YFP indicates fluorescence channel triggered by BiFC interaction. The DsRed indicates fluorescence channel for nuclear localization maker. Chl indicates the chloroplast spontaneous fluorescence channel. Bright indicates the bright field. Merge indicates channel combined the GFP,DsRed,Chl and the bright field. Scale bar = 10 μm
| [1] |
Wasternack C, Hause B. Jasmonates:biosynjournal, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany[J]. Ann Bot, 2013, 111(6):1021-1058.
doi: 10.1093/aob/mct067 URL |
| [2] |
Staswick PE, Su W, Howell SH. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant[J]. PNAS, 1992, 89(15):6837-6840.
pmid: 11607311 |
| [3] |
Berens ML, Berry HM, Mine A, et al. Evolution of hormone signaling networks in plant defense[J]. Annu Rev Phytopathol, 2017, 55:401-425.
doi: 10.1146/phyto.2017.55.issue-1 URL |
| [4] |
Huang H, Liu B, Liu L, et al. Jasmonate action in plant growth and development[J]. J Exp Bot, 2017, 68(6):1349-1359.
doi: 10.1093/jxb/erw495 pmid: 28158849 |
| [5] |
Dombrecht B, Xue GP, Sprague SJ, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis[J]. Plant Cell, 2007, 19(7):2225-2245.
pmid: 17616737 |
| [6] |
Chini A, Fonseca S, Fernández G, et al. The JAZ family of repressors is the missing link in jasmonate signalling[J]. Nature, 2007, 448(7154):666-671.
doi: 10.1038/nature06006 URL |
| [7] |
Yan J, Zhang C, Gu M, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor[J]. Plant Cell, 2009, 21(8):2220-2236.
doi: 10.1105/tpc.109.065730 URL |
| [8] |
Melotto M, Mecey C, Niu Y, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein[J]. Plant J, 2008, 55(6):979-988.
doi: 10.1111/tpj.2008.55.issue-6 URL |
| [9] |
Sheard LB, Tan X, Mao H, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor[J]. Nature, 2010, 468(7322):400-405.
doi: 10.1038/nature09430 URL |
| [10] |
Thines B, Katsir L, Melotto M, et al. JAZ repressor proteins are targets of the SCF(COI1)complex during jasmonate signalling[J]. Nature, 2007, 448(7154):661-665.
doi: 10.1038/nature05960 URL |
| [11] |
Zhang Z, Li X, Yu R, et al. Isolation, structural analysis, and expression characteristics of the maize TIFY gene family[J]. Mol Genet Genomics, 2015, 290(5):1849-1858.
doi: 10.1007/s00438-015-1042-6 URL |
| [12] |
Du M, Zhao J, Tzeng DTW, et al. MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato[J]. Plant Cell, 2017, 29(8):1883-1906.
doi: 10.1105/tpc.16.00953 URL |
| [13] |
Chen Q, Sun J, Zhai Q, et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis[J]. Plant Cell, 2011, 23(9):3335-3352.
doi: 10.1105/tpc.111.089870 URL |
| [14] |
Hong GJ, Xue XY, Mao YB, et al. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression[J]. Plant Cell, 2012, 24(6):2635-2648.
doi: 10.1105/tpc.112.098749 URL |
| [15] |
Fu J, Liu L, Liu Q, et al. ZmMYC2 exhibits diverse functions and enhances JA signaling in transgenic Arabidopsis[J]. Plant Cell Rep, 2020, 39(2):273-288.
doi: 10.1007/s00299-019-02490-2 URL |
| [16] |
Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses[J]. Plant Cell, 2011, 23(2):701-715.
doi: 10.1105/tpc.110.080788 URL |
| [17] |
Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis[J]. J Exp Bot, 2011, 62(6):2143-2154.
doi: 10.1093/jxb/erq408 URL |
| [18] |
Cheng Z, Sun L, Qi T, et al. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis[J]. Mol Plant, 2011, 4(2):279-288.
doi: 10.1093/mp/ssq073 URL |
| [19] |
Browse J, Wallis JG. Arabidopsis flowers unlocked the mechanism of jasmonate signaling[J]. Plants, 2019, 8(8):285.
doi: 10.3390/plants8080285 URL |
| [20] |
Wu H, Ye HY, Yao RF, et al. OsJAZ9 Acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice[J]. Plant Sci, 2015, 232:1-12.
doi: 10.1016/j.plantsci.2014.12.010 URL |
| [21] |
Acosta IF, Laparra H, Romero SP, et al. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize[J]. Science, 2009, 323(5911):262-265.
doi: 10.1126/science.1164645 pmid: 19131630 |
| [22] |
Xiao S, Dai LY, Liu FQ, et al. COS1:an Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence[J]. Plant Cell, 2004, 16(5):1132-1142.
pmid: 15075400 |
| [23] |
Shan XY, Wang JX, Chua L, et al. The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence[J]. Plant Physiol, 2011, 155(2):751-764.
doi: 10.1104/pp.110.166595 URL |
| [24] |
Feys B, Benedetti CE, Penfold CN, et al. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen[J]. Plant Cell, 1994, 6(5):751-759.
doi: 10.2307/3869877 URL |
| [25] |
Li L, Zhao Y, McCaig BC, et al. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development[J]. Plant Cell, 2004, 16(1):126-143.
doi: 10.1105/tpc.017954 URL |
| [26] |
晏胜伟, 孙程, 周晓今, 等. 玉米JAZ家族基因ZmJAZ4的克隆及功能分析[J]. 生物技术通报, 2015, 31(3):96-101.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.04.013 |
| Yan SW, Sun C, Zhou XJ, et al. Cloning and characterization analysis of zm JAZ4, a JAZ family gene in maize(Zea mays L.)[J]. Biotechnol Bull, 2015, 31(3):96-101. | |
| [27] |
Zheng J, Fu J, Gou M, et al. Genome-wide transcriptome analysis of two maize inbred lines under drought stress[J]. Plant Mol Biol, 2010, 72(4/5):407-421.
doi: 10.1007/s11103-009-9579-6 URL |
| [1] | YANG Hua-jie, ZHOU Yu-ping, FAN Tian, LV Tian-xiao, XIE Chu-ping, TIAN Chang-en. Screening and Identification of IQM4-Interacting Proteins in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2021, 37(11): 190-196. |
| [2] | WANG Yi-qiao, ZHAO Xin-jie, NIU Fang-fang, GUO Xiao-hua, YANG Bo, JIANG Yuan-qing. Analysis of Transcriptional Activity and Interaction Proteins of WRKY31 Transcription Factor in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2018, 34(5): 101-109. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||