








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (2): 141-149.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0241
Previous Articles Next Articles
YE Peng-lin( ), Kwasi Kyere-Yeboah, GAO E-bin(
), Kwasi Kyere-Yeboah, GAO E-bin( )
)
Received:2021-03-03
Online:2022-02-26
Published:2022-03-09
Contact:
GAO E-bin
E-mail:1776455793@qq.com;gaofei@ujs.edu.cn
YE Peng-lin, Kwasi Kyere-Yeboah, GAO E-bin. Effects on the Biosynthesis of Ethanol by Promoters PpetE and Pcpc560 in Synechocystis sp. PCC 6803[J]. Biotechnology Bulletin, 2022, 38(2): 141-149.
| 名称 Name | 特点 Characteristic |
|---|---|
| pMD18-T | 氨苄霉素/AmpR |
| pMD0168 | 氨苄霉素/AmpR 将slr0168的上下游600 bp基因扩增并克隆到质粒pMD18-T的SphI/MluI和XbaI/KpnI位点 |
| pBE406 | 壮观霉素/ SpR 将壮观霉素基因扩增并克隆到所构建质粒pMD0168的XhoI/XbaI位点 |
| pBE407 | 壮观霉素/ SpR 将启动子PpetE扩增并克隆到所构建质粒pBE406的BamHI/SalI位点,将终止子TrbcL扩增并克隆到所构建质粒pBE406的SalI/HindIII位点 |
| pBE407-pdc | 壮观霉素/ SpR pdc基因被扩增并和质粒pBE407双酶切,通过NdeI/BamHI位点相连接获得质粒pBE407-pdc |
| pBE01 | 壮观霉素/ SpR yqhD 基因被扩增并和质粒pBE407-pdc双酶切,通过XbaI/KpnI位点相连接获得质粒pBE01 |
| pBE408 | 壮观霉素/ SpR 将启动子Pcpc560扩增并克隆到所构建质粒pBE406的BamHI/SalI位点,将终止子TrbcL扩增并克隆到所构建质粒pBE406的SalI/HindIII位点 |
| pBE408-pdc | 壮观霉素/ SpR pdc基因被扩增并和质粒pBE408双酶切,通过NdeI/BamHI位点相连接获得质粒pBE408-pdc |
| pBE02 | 壮观霉素/ SpR yqhD 基因被扩增并和质粒pBE408-pdc双酶切,通过XbaI/KpnI位点相连接获得质粒pBE02 |
Table 1 Plasmids used in this study
| 名称 Name | 特点 Characteristic |
|---|---|
| pMD18-T | 氨苄霉素/AmpR |
| pMD0168 | 氨苄霉素/AmpR 将slr0168的上下游600 bp基因扩增并克隆到质粒pMD18-T的SphI/MluI和XbaI/KpnI位点 |
| pBE406 | 壮观霉素/ SpR 将壮观霉素基因扩增并克隆到所构建质粒pMD0168的XhoI/XbaI位点 |
| pBE407 | 壮观霉素/ SpR 将启动子PpetE扩增并克隆到所构建质粒pBE406的BamHI/SalI位点,将终止子TrbcL扩增并克隆到所构建质粒pBE406的SalI/HindIII位点 |
| pBE407-pdc | 壮观霉素/ SpR pdc基因被扩增并和质粒pBE407双酶切,通过NdeI/BamHI位点相连接获得质粒pBE407-pdc |
| pBE01 | 壮观霉素/ SpR yqhD 基因被扩增并和质粒pBE407-pdc双酶切,通过XbaI/KpnI位点相连接获得质粒pBE01 |
| pBE408 | 壮观霉素/ SpR 将启动子Pcpc560扩增并克隆到所构建质粒pBE406的BamHI/SalI位点,将终止子TrbcL扩增并克隆到所构建质粒pBE406的SalI/HindIII位点 |
| pBE408-pdc | 壮观霉素/ SpR pdc基因被扩增并和质粒pBE408双酶切,通过NdeI/BamHI位点相连接获得质粒pBE408-pdc |
| pBE02 | 壮观霉素/ SpR yqhD 基因被扩增并和质粒pBE408-pdc双酶切,通过XbaI/KpnI位点相连接获得质粒pBE02 |
| 引物序列Sequence of a primer | 大小Size |
|---|---|
| SP-F:5'-CCACGCGTAAGCTTGGATCCGCTCACGCAACTGGTCCAGAA-3' SP-R:5'-CGGGAGCTCGAATTCTAGAGTGCTTAGTGCATCTAACGC-3' | 1.1 kb |
| Pcpc-F:5'-CGTCTAGAGGATCCCCTGTAGAGAAGAGTCCCTG-3' Pcpc-R:5'-TTTCTCCTCTTTTGAATTAATCTCCTACTTGACTTTATGAG-3' | 550 bp |
| PetE-F:5'-GCTCTAGACAAGGATTCATAGCGGTTGCCCAATC-3' PetE-R:5'-GCTGCCTAGGATTCTGGCGAAAGGGGGATGTG-3' | 200 bp |
| TrbcL-F:5'-CGCGTCGACCGGTGTTTGGATTGTCGGAGT-3' TrbcL-R:5'-CCGACGCGTAAGCTTCCGGTAATTGGTAAATTGCTGTC-3' | 250 bp |
| Pdc-F:5'-CCGAGATCTCATATGTCCTACACCGTGGGCACCT-3' Pdc-R:5'-CGCGGATCCTGCAGCTCGAGTCTAGATTACAACAATTTGTTCACGGGT-3' | 1.7 kb |
| YqhD-F:5'-CAAACTCGAGTCTAGATGAACAACTTTAACTTGCACACCCCCAC-3' YqhD-R:5'-CGGGGTACCTGCAGTTAGCGGGCGGCTTCGTATATACGGC-3' | 1.2 kb |
| slr0168Up-F:5'-GGCATGCCGAGCGGCACCACGGGGCACCACCGC-3' slr0168Up-R:5'-GACGCGTCGGCGCACAGCAGCGTGCGACGTGTG-3' | 600 bp |
| slr0168Dw-F:5'-CTCTAGAGTGCCACTACCTGGCGTGCCGCTACC-3' slr0168Dw-R:5'-GGGGTACCCCGCATGACCAGCTGCCGCCCCAGC-3' | 600 bp |
Table 2 Primers used in this study
| 引物序列Sequence of a primer | 大小Size |
|---|---|
| SP-F:5'-CCACGCGTAAGCTTGGATCCGCTCACGCAACTGGTCCAGAA-3' SP-R:5'-CGGGAGCTCGAATTCTAGAGTGCTTAGTGCATCTAACGC-3' | 1.1 kb |
| Pcpc-F:5'-CGTCTAGAGGATCCCCTGTAGAGAAGAGTCCCTG-3' Pcpc-R:5'-TTTCTCCTCTTTTGAATTAATCTCCTACTTGACTTTATGAG-3' | 550 bp |
| PetE-F:5'-GCTCTAGACAAGGATTCATAGCGGTTGCCCAATC-3' PetE-R:5'-GCTGCCTAGGATTCTGGCGAAAGGGGGATGTG-3' | 200 bp |
| TrbcL-F:5'-CGCGTCGACCGGTGTTTGGATTGTCGGAGT-3' TrbcL-R:5'-CCGACGCGTAAGCTTCCGGTAATTGGTAAATTGCTGTC-3' | 250 bp |
| Pdc-F:5'-CCGAGATCTCATATGTCCTACACCGTGGGCACCT-3' Pdc-R:5'-CGCGGATCCTGCAGCTCGAGTCTAGATTACAACAATTTGTTCACGGGT-3' | 1.7 kb |
| YqhD-F:5'-CAAACTCGAGTCTAGATGAACAACTTTAACTTGCACACCCCCAC-3' YqhD-R:5'-CGGGGTACCTGCAGTTAGCGGGCGGCTTCGTATATACGGC-3' | 1.2 kb |
| slr0168Up-F:5'-GGCATGCCGAGCGGCACCACGGGGCACCACCGC-3' slr0168Up-R:5'-GACGCGTCGGCGCACAGCAGCGTGCGACGTGTG-3' | 600 bp |
| slr0168Dw-F:5'-CTCTAGAGTGCCACTACCTGGCGTGCCGCTACC-3' slr0168Dw-R:5'-GGGGTACCCCGCATGACCAGCTGCCGCCCCAGC-3' | 600 bp |
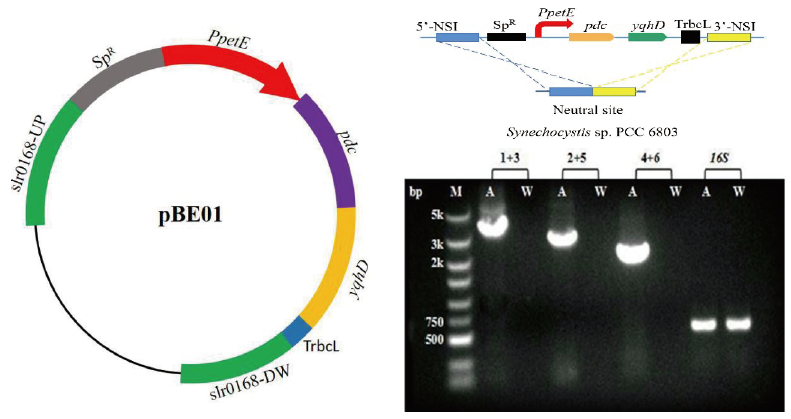
Fig.2 Construction diagram and PCR analysis of the SYN01 strain M:DNA marker. A:Extracted DNA from transformant genome. W:Extracted DNA from wild-type Synechocystis. 1:slr0168 upstream gene upstream primer slr0168Up-F. 2:pdc gene upstream primer Pdc-F. 3:pdc gene downstream primer Pdc-R. 4:yqhD gene upstream primer YqhD-F. 5:yqhD gene downstream primer YqhD-R. 6:slr0168 downstream gene downstream primer slr0168DW-R. 16S:Amplify the endogenous gene encoding 16S rRNA(600 bp). 1+3:Using primer 1 and primer 3 for amplification(3.6 kb). 2+5:Using primer 2 and primer 5 for amplification(2.9 kb). 4+6:Using primer 4 and primer 6 for amplification(2 kb)
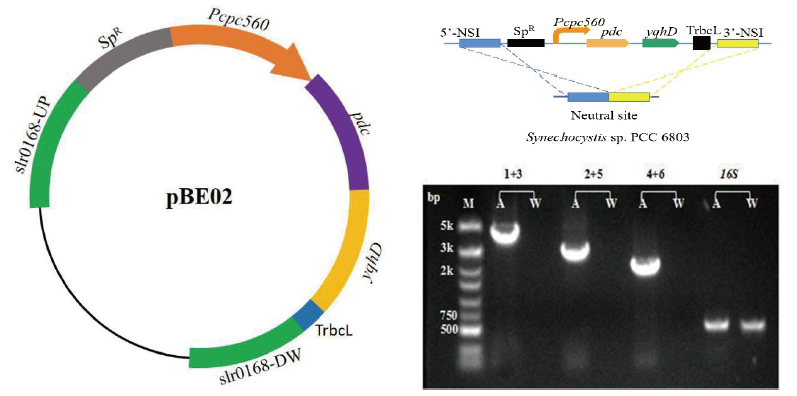
Fig.3 Construction diagram and PCR analysis of the SYN02 strain M:DNA marker. A:Extracted DNA from transformant genome. W:Extracted DNA from wild-type Synechocystis. 1:slr0168 upstream gene upstream primer slr0168Up-F. 2:pdc gene upstream primer Pdc-F. 3:pdc gene downstream primer Pdc-R. 4:yqhD gene upstream primer YqhD-F. 5:yqhD gene downstream primer YqhD-R. 6:slr0168 downstream gene downstream primer slr0168DW-R. 16S:Amplify the endogenous gene encoding 16S rRNA(600 bp). 1+3:Using primer 1 and primer 3 for amplification(3.9 kb). 2+5:Using primer 2 and primer 5 for amplification(2.9 kb). 4+6:Using primer 4 and primer 6 for amplification(2 kb)
| 基因Gene | 菌株Strain | 16S | CT1 | CT2 | CT3 | 平均 CT Average CT | ∆CT | 2-∆∆CT | 标准偏差SD |
|---|---|---|---|---|---|---|---|---|---|
| Pdc | SYN01 | 22.85 | 22.97 | 22.90 | 22.98 | 22.95 | 0.10 | 2.55 | 0.0252 |
| SYN02 | 22.85 | 21.60 | 21.57 | 21.63 | 21.60 | -1.25 | 0.0173 | ||
| YqhD | SYN01 | 22.85 | 23.76 | 23.72 | 24.04 | 23.84 | 0.99 | 2.31 | 0.1007 |
| SYN02 | 22.85 | 22.60 | 22.66 | 22.63 | 22.63 | -0.22 | 0.0173 |
Table 3 Reverse transcription of quantitative PCR results
| 基因Gene | 菌株Strain | 16S | CT1 | CT2 | CT3 | 平均 CT Average CT | ∆CT | 2-∆∆CT | 标准偏差SD |
|---|---|---|---|---|---|---|---|---|---|
| Pdc | SYN01 | 22.85 | 22.97 | 22.90 | 22.98 | 22.95 | 0.10 | 2.55 | 0.0252 |
| SYN02 | 22.85 | 21.60 | 21.57 | 21.63 | 21.60 | -1.25 | 0.0173 | ||
| YqhD | SYN01 | 22.85 | 23.76 | 23.72 | 24.04 | 23.84 | 0.99 | 2.31 | 0.1007 |
| SYN02 | 22.85 | 22.60 | 22.66 | 22.63 | 22.63 | -0.22 | 0.0173 |
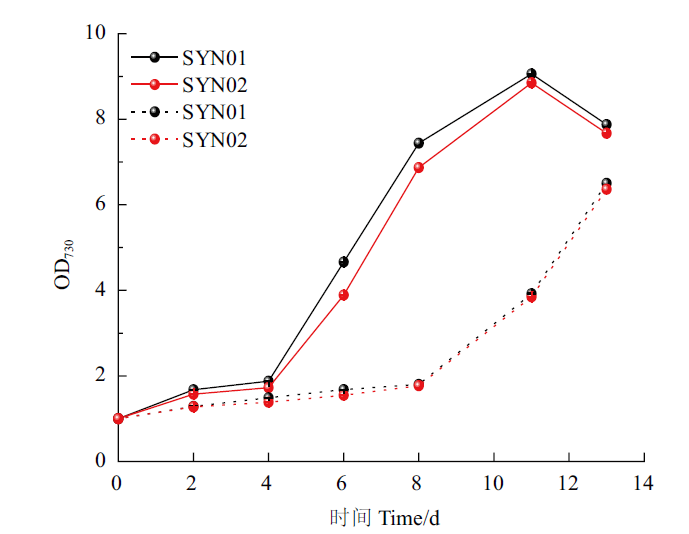
Fig.8 Growth condition of the strains under the optimized culture The solid line refers to 5% CO2+95% air,and the dashed line refers to 100% air. The same below
| [1] |
Song CF, Liu QL, Deng S, et al. Cryogenic-based CO2 capture technologies:State-of-the-art developments and current challenges[J]. Renew Sustain Energy Rev, 2019, 101:265-278.
doi: 10.1016/j.rser.2018.11.018 URL |
| [2] |
Afgan NH, Gobaisi DA, Carvalho MG, et al. Sustainable energy development[J]. Renew Sustain Energy Rev, 1998, 2(3):235-286.
doi: 10.1016/S1364-0321(98)00002-1 URL |
| [3] |
Nemeth N. Environment and energy:Problems, resolutions, solutions[J]. Int J Hydrog Energy, 1990, 15(7):457-462.
doi: 10.1016/0360-3199(90)90102-5 URL |
| [4] |
Goldemberg J . Special issue on sustainability and energy:Perspectives:“Ethanol for a sustainable energy future”[J]. Science, 2007, 317(5843):1325-1325.
doi: 10.1126/science.317.5843.1325 URL |
| [5] |
Parmar A, Singh NK, Pandey A, et al. Cyanobacteria and microalgae:a positive prospect for biofuels[J]. Bioresour Technol, 2011, 102(22):10163-10172.
doi: 10.1016/j.biortech.2011.08.030 URL |
| [6] |
Diao J, Song X, Zhang L, et al. Tailoring cyanobacteria as a new platform for highly efficient synjournal of astaxanthin[J]. Metab Eng, 2020, 61:275-287.
doi: 10.1016/j.ymben.2020.07.003 URL |
| [7] | 高宏, 唐蜻, 徐旭东. 集胞藻PCC6803铜离子诱导表达平台的构建[J]. 水生生物学报, 2007, 31(2):240-244. |
| Gao H, Tang Q, Xu XD. Construction of copper-induced gene expression platform in Synechocystis sp pcc6803[J]. Acta Hydrobiol Sin, 2007, 31(2):240-244. | |
| [8] |
Ghassemian M, Wong B, Ferreira F, et al. Cloning, sequencing and transcriptional studies of the genes for cytochrome c-553 and plastocyanin from Anabaena sp. PCC 7120[J]. Microbiology, 1994, 140(5):1151-1159.
doi: 10.1099/13500872-140-5-1151 URL |
| [9] |
Gao ZX, Zhao H, Li ZM, et al. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria[J]. Energy Environ Sci, 2012, 5(12):9857-9865.
doi: 10.1039/C2EE22675H URL |
| [10] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [11] |
Woo JE, Jang YS. Metabolic engineering of microorganisms for the production of ethanol and butanol from oxides of carbon[J]. Appl Microbiol Biotechnol, 2019, 103(20):8283-8292.
doi: 10.1007/s00253-019-10072-1 URL |
| [12] |
Namakoshi K, Nakajima T, Yoshikawa K, et al. Combinatorial deletions of glgC and PhaCE enhance ethanol production in Synechocystis sp. PCC 6803[J]. J Biotechnol, 2016, 239:13-19.
doi: 10.1016/j.jbiotec.2016.09.016 URL |
| [13] |
Mock M, Schmid A, Bühler K. Photoautotrophic production of succinate via the oxidative branch of the tricarboxylic acid cycle influences glycogen accumulation in Synechocystis sp. PCC 6803[J]. Algal Res, 2019, 43:101645.
doi: 10.1016/j.algal.2019.101645 URL |
| [14] | Noreña-Caro D, Benton MG. Cyanobacteria as photoautotrophic biofactories of high-value chemicals[J]. J CO2 Util, 2018, 28:335-366. |
| [15] |
Angermayr SA, Gorchs Rovira A, Hellingwerf KJ. Metabolic engineering of cyanobacteria for the synjournal of commodity products[J]. Trends Biotechnol, 2015, 33(6):352-361.
doi: 10.1016/j.tibtech.2015.03.009 pmid: 25908503 |
| [16] |
Yoshida S, Tanaka H, Hirayama M, et al. Production of pyruvate from mannitol by mannitol-assimilating pyruvate decarboxylase-negative Saccharomyces cerevisiae[J]. Bioengineered, 2015, 6(6):347-350.
doi: 10.1080/21655979.2015.1112472 pmid: 26588105 |
| [17] |
Takahashi H, Uchimiya H, Hihara Y. Difference in metabolite levels between photoautotrophic and photomixotrophic cultures of Synechocystis sp. PCC 6803 examined by capillary electrophoresis electrospray ionization mass spectrometry[J]. J Exp Bot, 2008, 59(11):3009-3018.
doi: 10.1093/jxb/ern157 pmid: 18611912 |
| [18] |
Miao R, Liu X, Englund E, et al. Isobutanol production in Synechocystis PCC 6803 using heterologous and endogenous alcohol dehydrogenases[J]. Metab Eng Commun, 2017, 5:45-53.
doi: 10.1016/j.meteno.2017.07.003 pmid: 29188183 |
| [19] |
Dexter J, Fu PC. Metabolic engineering of cyanobacteria for ethanol production[J]. Energy Environ Sci, 2009, 2(8):857.
doi: 10.1039/b811937f URL |
| [20] |
Yoshikawa K, Hirasawa T, Shimizu H. Effect of malic enzyme on ethanol production by Synechocystis sp. PCC 6803[J]. J Biosci Bioeng, 2015, 119(1):82-84.
doi: 10.1016/j.jbiosc.2014.06.001 URL |
| [21] |
Gao EB, Kyere-Yeboah K, Wu JH, et al. Photoautotrophic production of p-Coumaric acid using genetically engineered Synechocystis sp. Pasteur Culture Collection 6803[J]. Algal Res, 2021, 54:102180.
doi: 10.1016/j.algal.2020.102180 URL |
| [22] |
Zhou J, Zhang H, Meng H, et al. Discovery of a super-strong promoter enables efficient production of heterologous proteins in cyanobacteria[J]. Sci Rep, 2014, 4:4500.
doi: 10.1038/srep04500 pmid: 24675756 |
| [23] | Castenholz RW. Culturing methods for cyanobacteria[M]// Methods in Enzymology. Amsterdam:Elsevier, 1988:68-93. |
| [24] | Moss NA, Leao T, Glukhov E, et al. Collection, culturing, and genome analyses of tropical marine filamentous benthic cyanobacteria[M]// Methods in Enzymology. Amsterdam:Elsevier, 2018:3-43. |
| [25] |
Velmurugan R, Incharoensakdi A. Co-cultivation of two engineered strains of Synechocystis sp. PCC 6803 results in improved bioethanol production[J]. Renew Energy, 2020, 146:1124-1133.
doi: 10.1016/j.renene.2019.07.025 URL |
| [1] | LIU Yu-ling, WANG Meng-yao, SUN Qi, MA Li-hua, ZHU Xin-xia. Effect of RD29A Promoter on the Stress Resistance of Transgenic Tobacco with SikCDPK1 Gene from Saussurea involucrata [J]. Biotechnology Bulletin, 2023, 39(9): 168-175. |
| [2] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [3] | CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium [J]. Biotechnology Bulletin, 2023, 39(9): 84-96. |
| [4] | LIU Bao-cai, CHEN Jing-ying, ZHANG Wu-jun, HUANG Ying-zhen, ZHAO Yun-qing, LIU Jian-chao, WEI Zhi-cheng. Characteristics Analysis of Seed Microrhizome Gene Expression of Polygonatum cyrtonema [J]. Biotechnology Bulletin, 2023, 39(8): 220-233. |
| [5] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [6] | YE Yun-fang, TIAN Qing-yin, SHI Ting-ting, WANG Liang, YUE Yuan-zheng, YANG Xiu-lian, WANG Liang-gui. Research Progress in the Biosynthesis and Regulation of β-ionone in Plants [J]. Biotechnology Bulletin, 2023, 39(8): 91-105. |
| [7] | WANG Ling, ZHUO Shen, FU Xue-sen, LIU Zi-xuan, LIU Xiao-rong, WANG Zhi-hui, ZHOU Ri-bao, LIU Xiang-dan. Advances in the Biosynthetic Pathways and Related Genes of Lotus Alkaloids [J]. Biotechnology Bulletin, 2023, 39(7): 56-66. |
| [8] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [9] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [10] | ZHOU Ding-ding, LI Hui-hu, TANG Xing-yong, YU Fa-xin, KONG Dan-yu, LIU Yi. Research Progress in the Biosynthesis and Regulation of Glycyrrhizic Acid and Liquiritin [J]. Biotechnology Bulletin, 2023, 39(5): 44-53. |
| [11] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [12] | YU Hui-li, LI Ai-tao. Application of Cytochrome P450 in the Biosynthesis of Flavors and Fragrances [J]. Biotechnology Bulletin, 2023, 39(4): 24-37. |
| [13] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [14] | WANG Qi, HU Zhe, FU Wei, LI Guang-zhe, HAO Lin. Regulation of Burkholderia sp. GD17 on the Drought Tolerance of Cucumber Seedlings [J]. Biotechnology Bulletin, 2023, 39(3): 163-175. |
| [15] | YAO Xiao-wen, LIANG Xiao, CHEN Qing, WU Chun-ling, LIU Ying, LIU Xiao-qiang, SHUI Jun, QIAO Yang, MAO Yi-ming, CHEN Yin-hua, ZHANG Yin-dong. Study on the Expression Pattern of Genes in Lignin Biosynthesis Pathway of Cassava Resisting to Tetranychus urticae [J]. Biotechnology Bulletin, 2023, 39(2): 161-171. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||