








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (4): 117-125.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0904
Previous Articles Next Articles
GAO Meng1( ), LI Fu-ting1, WEI Zhan-lin1, ZHANG Sai-hang2, BAI Ru-qian1, SHANG Yi1, MA Ling1(
), LI Fu-ting1, WEI Zhan-lin1, ZHANG Sai-hang2, BAI Ru-qian1, SHANG Yi1, MA Ling1( )
)
Received:2021-10-21
Online:2022-04-26
Published:2022-05-06
Contact:
MA Ling
E-mail:simone_gao@126.com;maling223322@126.com
GAO Meng, LI Fu-ting, WEI Zhan-lin, ZHANG Sai-hang, BAI Ru-qian, SHANG Yi, MA Ling. Component Analysis of SCFSLF Complex in Diploid Potato[J]. Biotechnology Bulletin, 2022, 38(4): 117-125.
| 引物名称 Primer | 引物序列 Sequence(5'-3') |
|---|---|
| 4330-SSK1-F | ATGGAGTTGTGTTGCCAATC |
| 4330-SSK1-R | AATCTCAGGACCTTCAAATGC |
| 1040-CLU1-F | GTGTCATGTGCCAAGTACAA |
| 1040-CLU1-R | CATAACGTCTATCCTTGCCG |
| QEF1a-F | GATGGTCAGACCCGTGAACATGC |
| QEF1a-R | ACGATTTCATCATACCTAGCCTTGGAGT |
Table 1 Fluorescent quantitative primers for SSK1 and Cullin1 in potato(S. tuberosum)
| 引物名称 Primer | 引物序列 Sequence(5'-3') |
|---|---|
| 4330-SSK1-F | ATGGAGTTGTGTTGCCAATC |
| 4330-SSK1-R | AATCTCAGGACCTTCAAATGC |
| 1040-CLU1-F | GTGTCATGTGCCAAGTACAA |
| 1040-CLU1-R | CATAACGTCTATCCTTGCCG |
| QEF1a-F | GATGGTCAGACCCGTGAACATGC |
| QEF1a-R | ACGATTTCATCATACCTAGCCTTGGAGT |
| 基因编号Gene ID | 基因位点Location | E值E value | |
|---|---|---|---|
| PiSSK1同源基因 PiSSK1-homologous gene | Soltu.DM.11G004330 | chr11:4294163..4295355 reverse | 1.71E-102 |
| Soltu.DM.11G016290 | chr11:30776770..30778841 reverse | 2.83E-44 | |
| Soltu.DM.11G004300 | chr11:4286144..4286630 forward | 9.28E-43 | |
| Soltu.DM.10G017580 | chr10:47907512..47910644 forward | 9.30E-42 | |
| Soltu.DM.01G051200 | chr01:87717008..87719187 forward | 2.54E-41 | |
| Soltu.DM.11G004500 | chr11:4499656..4500133 reverse | 1.32E-40 | |
| PiCUL1-P同源基因 PiCUL1-P-homologous genes | Soltu.DM.06G001040 | chr06:1465504..1471494 reverse | 0 |
| Soltu.DM.06G035050 | chr06:58938032..58944924 reverse | 0 | |
| Soltu.DM.09G023090 | chr09:58921771..58929345 reverse | 0 | |
| Soltu.DM.01G022080 | chr01:60076281..60087621 forward | 0 | |
| Soltu.DM.09G003030 | chr09:2388523..2390740 reverse | 0 | |
| Soltu.DM.09G002940 | chr09:2349588..2351796 forward | 0 | |
| Soltu.DM.01G022100 | chr01:60106453..60114114 forward | 0 | |
| Soltu.DM.01G022150 | chr01:60164455..60171592 forward | 0 | |
| Soltu.DM.09G003010 | chr09:2377567..2379367 reverse | 0 | |
| Soltu.DM.01G022120 | chr01:60134989..60155075 forward | 0 | |
| PiRbx1同源基因 PiRbx1-homologous genes | Soltu.DM.06G022320 | chr06:48782308..48785375 reverse | 1.82E-71 |
Table 2 Homologous comparison of SCFSLF complex components in diploid potato DM
| 基因编号Gene ID | 基因位点Location | E值E value | |
|---|---|---|---|
| PiSSK1同源基因 PiSSK1-homologous gene | Soltu.DM.11G004330 | chr11:4294163..4295355 reverse | 1.71E-102 |
| Soltu.DM.11G016290 | chr11:30776770..30778841 reverse | 2.83E-44 | |
| Soltu.DM.11G004300 | chr11:4286144..4286630 forward | 9.28E-43 | |
| Soltu.DM.10G017580 | chr10:47907512..47910644 forward | 9.30E-42 | |
| Soltu.DM.01G051200 | chr01:87717008..87719187 forward | 2.54E-41 | |
| Soltu.DM.11G004500 | chr11:4499656..4500133 reverse | 1.32E-40 | |
| PiCUL1-P同源基因 PiCUL1-P-homologous genes | Soltu.DM.06G001040 | chr06:1465504..1471494 reverse | 0 |
| Soltu.DM.06G035050 | chr06:58938032..58944924 reverse | 0 | |
| Soltu.DM.09G023090 | chr09:58921771..58929345 reverse | 0 | |
| Soltu.DM.01G022080 | chr01:60076281..60087621 forward | 0 | |
| Soltu.DM.09G003030 | chr09:2388523..2390740 reverse | 0 | |
| Soltu.DM.09G002940 | chr09:2349588..2351796 forward | 0 | |
| Soltu.DM.01G022100 | chr01:60106453..60114114 forward | 0 | |
| Soltu.DM.01G022150 | chr01:60164455..60171592 forward | 0 | |
| Soltu.DM.09G003010 | chr09:2377567..2379367 reverse | 0 | |
| Soltu.DM.01G022120 | chr01:60134989..60155075 forward | 0 | |
| PiRbx1同源基因 PiRbx1-homologous genes | Soltu.DM.06G022320 | chr06:48782308..48785375 reverse | 1.82E-71 |
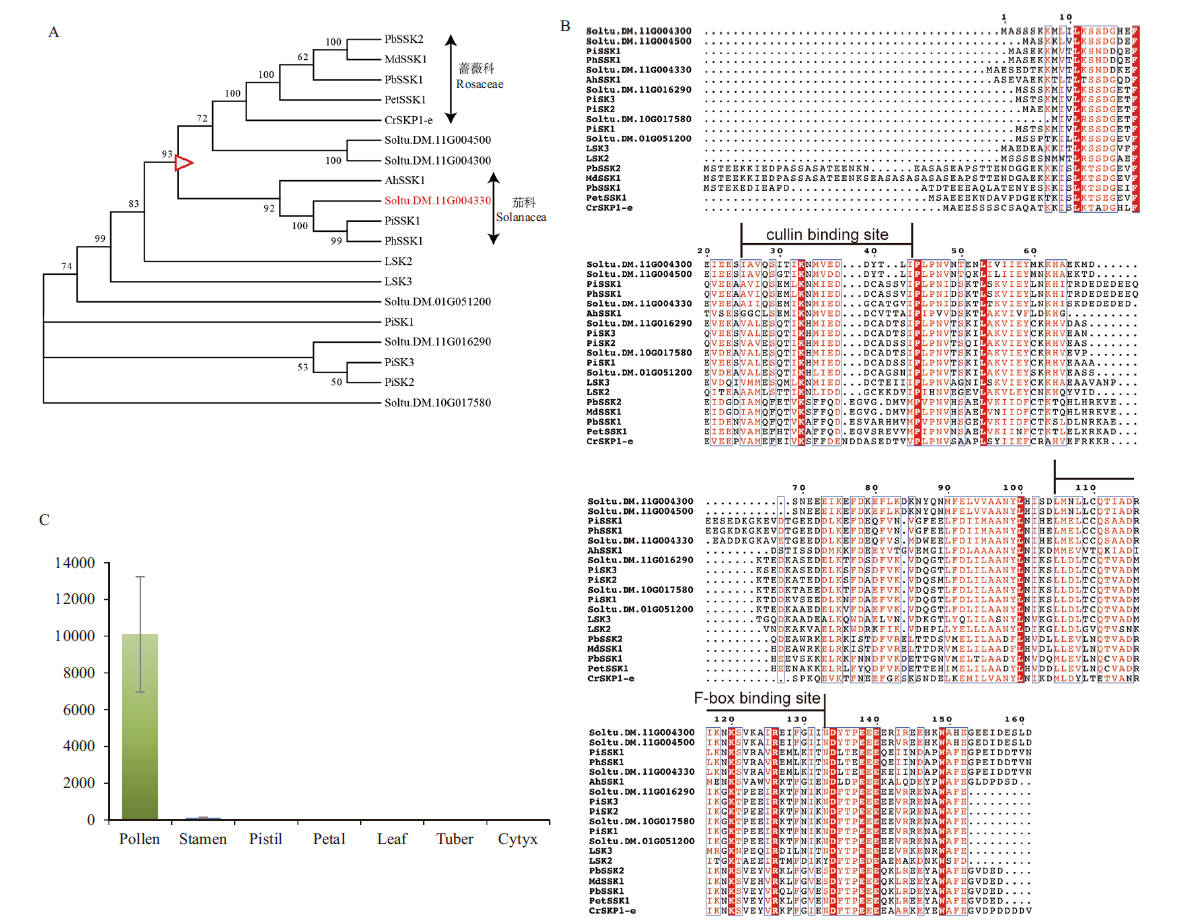
Fig. 2 SSK1 protein sequence and expression analysis A:Phylogenetic tree of SSK genes in GSI plants(The branch marked by the red triangle is the branch where Rosaceae and Solanaceae separated). B:Sequence alignment of SSK protein in GSI plants. C:The expression of Soltu.DM.11G004330 in different organs of E172 is calculated by 2-∆∆Ct with stamen as control
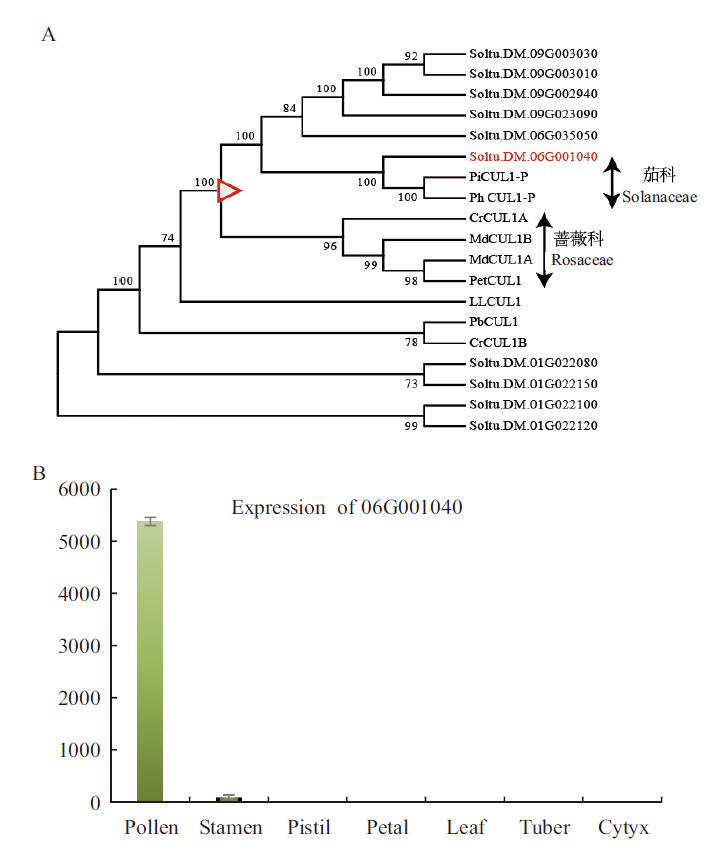
Fig. 3 Cullin1 protein evolution and expression analysis A:Phylogenetic tree of CUL genes in GSI plants(The branch marked by the red triangle is the branch where Rosaceae and Solanaceae separated). B:The expression of Soltu.DM.06G001040 in different organs of E172 is calculated by 2-∆∆Ct with stamen as control
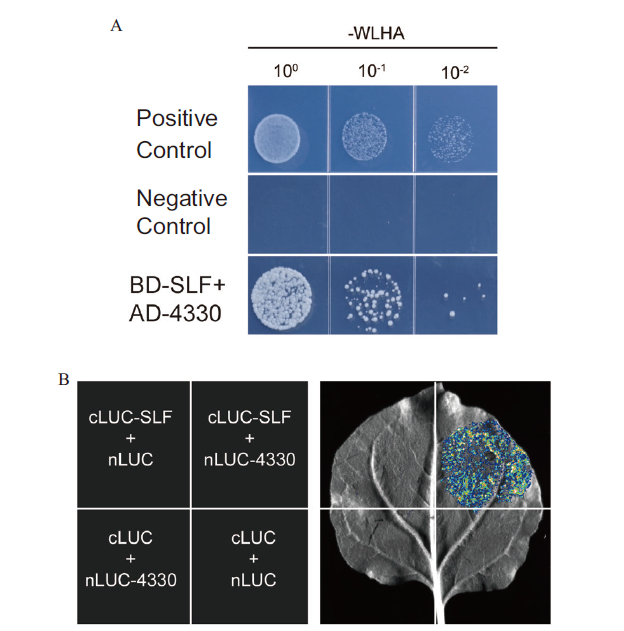
Fig. 5 Interaction analysis of Soltu.DM.11G004330 and SLF A:Y2H of Soltu.DM.11G004330 and SLF(-WLHA,synthetic dropout media lacking tryptophan,leucine,histidine,and adenine(-Trp/W,-Leu/L,-His/H,-adenine/Ade);100,10-1,and 10-2 indicates concentration of E.coli;pGBKT7-53 and pGADT7-T are used as positive control;pGBKT7-Lam and pGADT7-T are used as negative control;4330 is abbreviated from Soltu.DM.11G004330). B:LUC complementation of Soltu.DM.11G004330 and SLF.(The left figure denotes how injections were carried out on tobacco leaves and 4330 was the abbreviation of Soltu.DM.11G004330)
| [1] |
McClure B, Cruz-García F, Romero C. Compatibility and incompatibility in S-RNase-based systems[J]. Ann Bot, 2011, 108(4):647-658.
doi: 10.1093/aob/mcr179 URL |
| [2] |
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility[J]. Nat Plants, 2016, 2(9):16130.
doi: 10.1038/nplants.2016.130 pmid: 27595657 |
| [3] |
Matsumoto D, Tao R. Recognition of S-RNases by an S locus F-box like protein and an S haplotype-specific F-box like protein in the Prunus-specific self-incompatibility system[J]. Plant Mol Biol, 2019, 100(4/5):367-378.
doi: 10.1007/s11103-019-00860-8 URL |
| [4] |
Hua Z, Kao TH. Identification and characterization of components of a putative Petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility[J]. Plant Cell, 2006, 18(10):2531-2553.
doi: 10.1105/tpc.106.041061 URL |
| [5] |
Li S, Sun P, Williams JS, et al. Identification of the self-incompatibility locus F-box protein-containing complex in Petunia inflata[J]. Plant Reprod, 2014, 27(1):31-45.
doi: 10.1007/s00497-013-0238-3 URL |
| [6] | 李红艳, 翟赛亚, 徐明磊. 植物配子体型自交不亲和性作用机理研究进展[J]. 南方园艺, 2015, 26(3):76-78. |
| Li HY, Zhai SY, Xu ML. Research progress on gametophytic self-incompatibility in plants[J]. South Hortic, 2015, 26(3):76-78. | |
| [7] |
Williams JS, Wu L, Li S, et al. Insight into S-RNase-based self-incompatibility in Petunia:recent findings and future directions[J]. Front Plant Sci, 2015, 6:41.
doi: 10.3389/fpls.2015.00041 pmid: 25699069 |
| [8] |
Luu DT, Qin X, Morse D, et al. S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility[J]. Nature, 2000, 407(6804):649-651.
doi: 10.1038/35036623 URL |
| [9] |
Goldraij A, Kondo K, Lee CB, et al. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana[J]. Nature, 2006, 439(7078):805-810.
doi: 10.1038/nature04491 URL |
| [22] |
Kubo K, Entani T, Takara A, et al. Collaborative non-self recognition system in S-RNase-based self-incompatibility[J]. Science, 2010, 330(6005):796-799.
doi: 10.1126/science.1195243 URL |
| [23] |
Entani T, Kubo K, Isogai S, et al. Ubiquitin-proteasome-mediated degradation of S-RNase in a solanaceous cross-compatibility reaction[J]. Plant J, 2014, 78(6):1014-1021.
doi: 10.1111/tpj.12528 URL |
| [24] |
Kubo KI, Tsukahara M, Fujii S, et al. Cullin1-P is an essential component of non-self recognition system in self-incompatibility in Petunia[J]. Plant Cell Physiol, 2016, 57(11):2403-2416.
doi: 10.1093/pcp/pcw152 URL |
| [25] |
Chang LC, Guo CL, Lin YS, et al. Pollen-specific SKP1-like proteins are components of functional scf complexes and essential for lily pollen tube elongation[J]. Plant Cell Physiol, 2009, 50(8):1558-1572.
doi: 10.1093/pcp/pcp100 URL |
| [26] | Ren Y, Hua QZ, Pan JY, et al. SKP1-like protein, CrSKP1-e, interacts with pollen-specific F-box proteins and assembles into SCF-type E3 complex in ‘Wuzishatangju’(Citrus reticulata Blanco)pollen[J]. PeerJ, 2020, 8:e10578. |
| [27] |
Zeng B, Wang JY, Hao Q, et al. Identification of a novel SBP1-containing SCFSFB complex in wild dwarf almond(Prunus tenella)[J]. Front Genet, 2019, 10:1019. DOI: 10.3389/fgene.2019.01019.
doi: 10.3389/fgene.2019.01019 pmid: 31708966 |
| [28] |
Yuan H, Meng D, Gu ZY, et al. A novel gene, MdSSK1, as a component of the SCF complex rather than MdSBP1 can mediate the ubiquitination of S-RNase in apple[J]. J Exp Bot, 2014, 65(12):3121-3131.
doi: 10.1093/jxb/eru164 pmid: 24759884 |
| [29] |
Huang J, Zhao L, Yang QY, et al. AhSSK1, a novel SKP1-like protein that interacts with the S-locus F-box protein SLF[J]. Plant J, 2006, 46(5):780-793.
pmid: 16709194 |
| [30] |
Zhao L, Huang J, Zhao ZH, et al. The Skp1-like protein SSK1 is required for cross-pollen compatibility inS-RNase-based self-incompatibility[J]. Plant J, 2010, 62(1):52-63.
doi: 10.1111/j.1365-313X.2010.04123.x URL |
| [10] |
Roldán JA, Rojas HJ, Goldraij A. Disorganization of F-actin cytoskeleton precedes vacuolar disruption in pollen tubes during the in vivo self-incompatibility response in Nicotiana alata[J]. Ann Bot, 2012, 110(4):787-795.
doi: 10.1093/aob/mcs153 URL |
| [11] |
O’Brien M, Major G, Chantha SC, et al. Isolation of S-RNase binding proteins from Solanum chacoense:identification of an SBP1(RING finger protein)orthologue[J]. Sex Plant Reprod, 2004, 17(2):81-87.
doi: 10.1007/s00497-004-0218-8 URL |
| [12] |
McClure BA, Gray JE, Anderson MA, et al. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA[J]. Nature, 1990, 347(6295):757-760.
doi: 10.1038/347757a0 URL |
| [13] |
Xu C, Li MF, Wu JK, et al. Identification of a canonical scf(slf)complex involved In S-RNase-based Self-incompatibility Of Pyrus(rosaceae)[J]. Plant Mol Biol, 2013, 81(3):245-257.
doi: 10.1007/s11103-012-9995-x URL |
| [14] | 曾斌, 余镇藩, 马鑫鑫, 等. 基于S-核酸酶的自交不亲和cullin1基因的研究进展[J]. 分子植物育种, 2019, 17(2):466-470. |
| Zeng B, Yu ZF, Ma XX, et al. Research progress of self-incompatibility Cullin1 gene based on S-RNase[J]. Mol Plant Breed, 2019, 17(2):466-470. | |
| [15] |
Zheng N, Schulman BA, Song LZ, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex[J]. Nature, 2002, 416(6882):703-709.
doi: 10.1038/416703a URL |
| [16] |
Farras R. SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase[J]. EMBO J, 2001, 20(11):2742-2756.
doi: 10.1093/emboj/20.11.2742 URL |
| [17] | 秘彩莉, 刘旭, 张学勇. F-box蛋白质在植物生长发育中的功能[J]. 遗传, 2006, 28(10):1337-1342. |
| Bi CL, Liu X, Zhang XY. The function of F-box protein in plant growth and development[J]. Hereditas, 2006, 28(10):1337-1342. | |
| [18] |
Yu H, Zhang Y, Moss BL, et al. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response[J]. Nat Plants, 2015, 1:14030.
doi: 10.1038/nplants.2014.30 URL |
| [19] |
Qiao H, Wang F, Zhao L, et al. The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility[J]. Plant Cell, 2004, 16(9):2307-2322.
doi: 10.1105/tpc.104.024919 URL |
| [20] |
Entani T, Takayama S, Iwano M, et al. Relationship between polyploidy and pollen self-incompatibility phenotype in Petunia hybrida Vilm[J]. Biosci Biotechnol Biochem, 1999, 63(11):1882-1888.
doi: 10.1271/bbb.63.1882 URL |
| [21] |
Ushijima K, Sassa H, Dandekar AM, et al. Structural and transcriptional analysis of the self-incompatibility locus of almond:identification of a pollen-expressed F-box gene with haplotype-specific polymorphism[J]. Plant Cell, 2003, 15(3):771-781.
doi: 10.1105/tpc.009290 URL |
| [1] | LEI Chun-xia, LI Can-hui, CHEN Yong-kun, GONG Ming. Physiological and Biochemical Basis and Molecular Mechanism of Solanum tuberosum Tuberization [J]. Biotechnology Bulletin, 2022, 38(4): 44-57. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||