








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (4): 97-105.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1344
Previous Articles Next Articles
Olalekan Amoo( ), HU Li-min, ZHAI Yun-gu, FAN Chu-chuan(
), HU Li-min, ZHAI Yun-gu, FAN Chu-chuan( ), ZHOU Yong-ming
), ZHOU Yong-ming
Received:2021-10-25
Online:2022-04-26
Published:2022-05-06
Contact:
FAN Chu-chuan
E-mail:olalekan@webmail.hzau.edu.cn;fanchuchuan@mail.hzau.edu.cn
Olalekan Amoo, HU Li-min, ZHAI Yun-gu, FAN Chu-chuan, ZHOU Yong-ming. Regulation of Shoot Branching by BRANCHED1 in Brassica napus Based on Gene Editing Technology[J]. Biotechnology Bulletin, 2022, 38(4): 97-105.
| 引物 Primer | 序列 Sequence(5'-3') | 用途 Purpose |
|---|---|---|
| CBnaC01-F1 | GAGACCATAACCACACACAAC | Cloning BnaC01g34090D |
| CBnaC01-F2 | TGTGGACAGTCGAGGATAGA | |
| CBnaC01-R | TCCATGTAGACAGAGACGATTT | |
| CBnaA01-F1 | GAGACCATAACCACACACAAC | Cloning BnaA01g26700D |
| CBnaA01-F2 | TGTGGACAGTCGAGGATAGA | |
| CBnaA01-R | GACGATTGCCCACAAAGTAAA | |
| CBnaC05-F | CAGTACTACCACCACCATCAAT | Cloning BnaC05g51920D |
| CBnaC05-R | TCGGGATCTGAGATAGCCTATAA | |
| CBnaCnn-F | ACAACAACAACAGAAGATCTCTCAG | Cloning BnaCnng23770D |
| CBnaCnn-R | CATGAGGTCTCTTGGTCTCTCC | |
| CBnaA03-F1 | ACAACAGGTCTTTCAGTACTACC | Cloning BnaA03g34820D |
| CBnaA03-F2 | CAAGGACCGGCGTTAGG | |
| CBnaA03-R | CATGAGGTCTCTTGGTCTCTC | |
| BnBRC1T1-F | gtcAAGCATCATGCAGGAACATG | BnaBRC1 target construction |
| BnBRC1T1-R | aaacCATGTTCCTGCATGATGCT | |
| BnBRC1T2-F | attGCAAAGGGTCAGGTCCTGAA | |
| BnBRC1T2-R | aaacTTCAGGACCTGACCCTTTG | |
| BnBRC1T3-F | gtcACGGAGAGATTCTCACTCCCA | |
| BnBRC1T3-R | aaacTGGGAGTGAGAATCTCTCCG | |
| BnBRC1T4-F | gtcATGGATATCTCCGTAACTCTC | |
| BnBRC1T4-R | aaacGAGAGTTACGGAGATATCCA | |
| BnBRC1T5-F | attGTCGCCATGTCGAGGACCTCT | |
| BnBRC1T5-R | aaacAGAGGTCCTCGACATGGCGA | |
| BnBRC1T6-F | attGAGGAGATGGTCCATGTCAG | |
| BnBRC1T6-R | aaacCTGACATGGACCATCTCCT | |
| PB-L | GCGCGCgGTctcGCTCGACTAGTATGG | Transgenic positive detection |
| PB-R | GCGCGCggtctcTACCGACGCGTATCC | |
| PSH 20A-1F | ggagtgagtacggtgtgcGAGACCATAACCACACACAAC | BnaBRC1 editing check |
| PSH 20A-1R | gagttggatgctggatggCTCATTCTACGATCTCTTGTCCC | |
| PSH 20A-2F | ggagtgagtacggtgtgcGGGACAAGAGATCGTAGAATGAG | |
| PSH 20A-2R | gagttggatgctggatggATGTATACGTACGGGTTTGGG | |
| PSH 20A-3F | ggagtgagtacggtgtgcACACACATAGAAGATTCCCAGAG | |
| PSH 20A-3R | gagttggatgctggatggAGGCTGTTCGCGATCTTTAT | |
| PSH 20A-4F | ggagtgagtacggtgtgcCCTTTTTCTCACTTCGAATCCG | |
| PSH 20A-4R | gagttggatgctggatggTTGGTTTCTGAGGGCTCAAT | |
| PSH 20B-1F | ggagtgagtacggtgtgcTCTTACAAGCACCTTCTTCTTTTTC | |
| PSH 20B-1R | gagttggatgctggatggCAAGGGCTCAACGAGAGATT | |
| PSH 20B-2F | ggagtgagtacggtgtgcTTGACCACCACCATCATCAG | |
| PSH 20B-2R | gagttggatgctggatggGGGCTCAATGAGGTGAGATT | |
| PSH 20B-CF | ggagtgagtacggtgtgcCGGCACAGCAAGATCAAAAC | |
| PSH 20B-CR | gagttggatgctggatggGCTTGTGTGAGCAACCATTC |
Table 1 Primers used in this study
| 引物 Primer | 序列 Sequence(5'-3') | 用途 Purpose |
|---|---|---|
| CBnaC01-F1 | GAGACCATAACCACACACAAC | Cloning BnaC01g34090D |
| CBnaC01-F2 | TGTGGACAGTCGAGGATAGA | |
| CBnaC01-R | TCCATGTAGACAGAGACGATTT | |
| CBnaA01-F1 | GAGACCATAACCACACACAAC | Cloning BnaA01g26700D |
| CBnaA01-F2 | TGTGGACAGTCGAGGATAGA | |
| CBnaA01-R | GACGATTGCCCACAAAGTAAA | |
| CBnaC05-F | CAGTACTACCACCACCATCAAT | Cloning BnaC05g51920D |
| CBnaC05-R | TCGGGATCTGAGATAGCCTATAA | |
| CBnaCnn-F | ACAACAACAACAGAAGATCTCTCAG | Cloning BnaCnng23770D |
| CBnaCnn-R | CATGAGGTCTCTTGGTCTCTCC | |
| CBnaA03-F1 | ACAACAGGTCTTTCAGTACTACC | Cloning BnaA03g34820D |
| CBnaA03-F2 | CAAGGACCGGCGTTAGG | |
| CBnaA03-R | CATGAGGTCTCTTGGTCTCTC | |
| BnBRC1T1-F | gtcAAGCATCATGCAGGAACATG | BnaBRC1 target construction |
| BnBRC1T1-R | aaacCATGTTCCTGCATGATGCT | |
| BnBRC1T2-F | attGCAAAGGGTCAGGTCCTGAA | |
| BnBRC1T2-R | aaacTTCAGGACCTGACCCTTTG | |
| BnBRC1T3-F | gtcACGGAGAGATTCTCACTCCCA | |
| BnBRC1T3-R | aaacTGGGAGTGAGAATCTCTCCG | |
| BnBRC1T4-F | gtcATGGATATCTCCGTAACTCTC | |
| BnBRC1T4-R | aaacGAGAGTTACGGAGATATCCA | |
| BnBRC1T5-F | attGTCGCCATGTCGAGGACCTCT | |
| BnBRC1T5-R | aaacAGAGGTCCTCGACATGGCGA | |
| BnBRC1T6-F | attGAGGAGATGGTCCATGTCAG | |
| BnBRC1T6-R | aaacCTGACATGGACCATCTCCT | |
| PB-L | GCGCGCgGTctcGCTCGACTAGTATGG | Transgenic positive detection |
| PB-R | GCGCGCggtctcTACCGACGCGTATCC | |
| PSH 20A-1F | ggagtgagtacggtgtgcGAGACCATAACCACACACAAC | BnaBRC1 editing check |
| PSH 20A-1R | gagttggatgctggatggCTCATTCTACGATCTCTTGTCCC | |
| PSH 20A-2F | ggagtgagtacggtgtgcGGGACAAGAGATCGTAGAATGAG | |
| PSH 20A-2R | gagttggatgctggatggATGTATACGTACGGGTTTGGG | |
| PSH 20A-3F | ggagtgagtacggtgtgcACACACATAGAAGATTCCCAGAG | |
| PSH 20A-3R | gagttggatgctggatggAGGCTGTTCGCGATCTTTAT | |
| PSH 20A-4F | ggagtgagtacggtgtgcCCTTTTTCTCACTTCGAATCCG | |
| PSH 20A-4R | gagttggatgctggatggTTGGTTTCTGAGGGCTCAAT | |
| PSH 20B-1F | ggagtgagtacggtgtgcTCTTACAAGCACCTTCTTCTTTTTC | |
| PSH 20B-1R | gagttggatgctggatggCAAGGGCTCAACGAGAGATT | |
| PSH 20B-2F | ggagtgagtacggtgtgcTTGACCACCACCATCATCAG | |
| PSH 20B-2R | gagttggatgctggatggGGGCTCAATGAGGTGAGATT | |
| PSH 20B-CF | ggagtgagtacggtgtgcCGGCACAGCAAGATCAAAAC | |
| PSH 20B-CR | gagttggatgctggatggGCTTGTGTGAGCAACCATTC |

Fig.2 Motif analysis and conserved domain structures of BnaBRC1 gene A:Analysis of conserved motifs in the protein sequence of homologous copies of the BRC1 gene in B. napus,and A. thaliana. B:Prediction of the conserved domains in the homologous copies of BRC1 in B. napus and A. thaliana
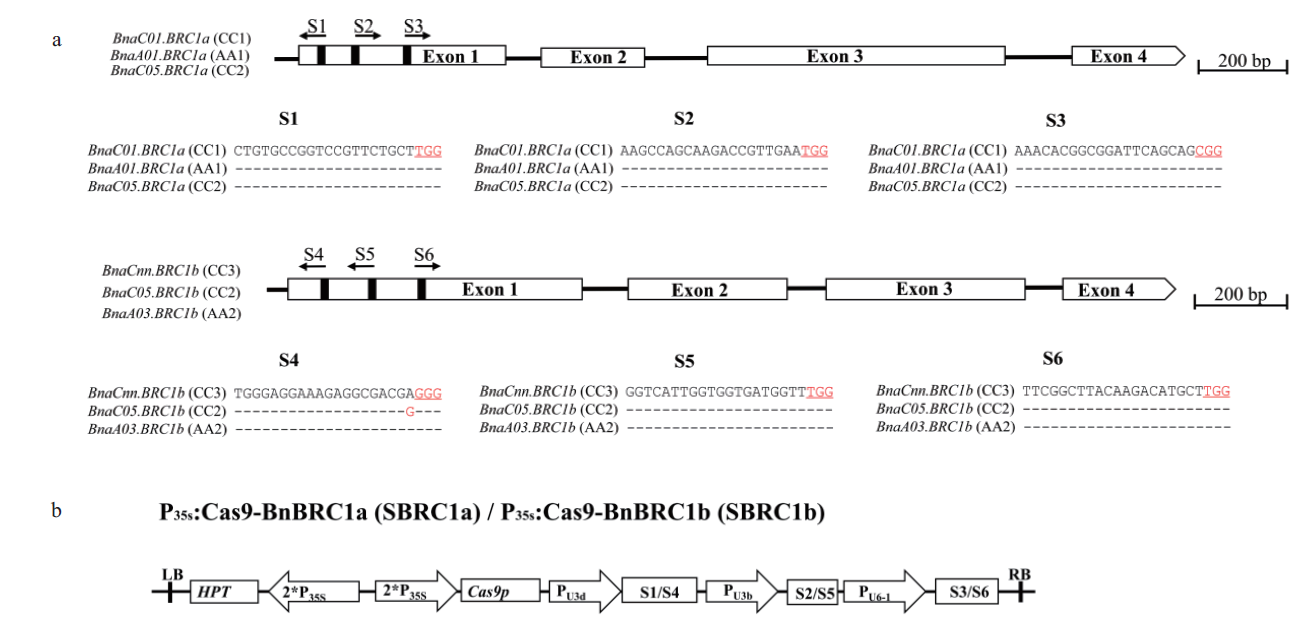
Fig.4 BnaBRC1 gene structure with target sequences and schematics of binary plasmid vectors a:The BnaBRC1 gene structure includes 4 exons separated by three introns. The vertical line in the gene structure indicates the target site,and the arrow indicates the sgRNA direction. The target sequences are shown with the PAM highlighted in red and underlined. b:Schematic presentation of binary vector SBRC1a and SBRC1b.

Fig.5 Mutated alleles at the S2 and S3 target sites of BnaBRC1 mutants from T0 to T1 generation CRISPR/Cas9-induced insertions and deletions are indicated by red font and red hyphens respectively,while the PAM is underlined and highlighted in red. “A” and “C” are the wild type alleles of the target gene on A and C chromosome copy,and “a”and “c” are the mutated alleles of the target gene on A and C chromosome copy. “_” and “ +” indicate the deletion and insertion of bases,respectively

Fig.6 Phenotypes of BnaBRC1 mutants A and B:Statistical analysis of leaf numbers and plant height(cm)of BnaBRC1 mutants,respectively. C and D:Phenotypes at the vegetative and flowering stage,respectively,WT refers to wild type(AA1CC1CC2),aacc1CC2 refers to double copy of homozygous BnaBRC1 mutant. The data and error bars represent the mean ± SD,n ≥ 10,and bar = 3 cm
| 靶点 On-target site | 基因 Gene | 测序植株数目 Number of sequenced plants | 脱靶位点数 Putative off-target site | 脱靶编辑 Off-target editing |
|---|---|---|---|---|
| S1 | BnaBRC1 | 72 | 9 | No |
| S2 | BnaBRC1 | 72 | 14 | No |
| S3 | BnaBRC1 | 72 | 11 | No |
| S4 | BnaBRC1 | 54 | 18 | No |
| S5 | BnaBRC1 | 54 | 18 | No |
| S6 | BnaBRC1 | 54 | 16 | No |
Table 2 Detection of potential off-target effects for each sgRNA target site in T0 mutated plants
| 靶点 On-target site | 基因 Gene | 测序植株数目 Number of sequenced plants | 脱靶位点数 Putative off-target site | 脱靶编辑 Off-target editing |
|---|---|---|---|---|
| S1 | BnaBRC1 | 72 | 9 | No |
| S2 | BnaBRC1 | 72 | 14 | No |
| S3 | BnaBRC1 | 72 | 11 | No |
| S4 | BnaBRC1 | 54 | 18 | No |
| S5 | BnaBRC1 | 54 | 18 | No |
| S6 | BnaBRC1 | 54 | 16 | No |
| [1] |
Jiang HF, Egli DB. Shade induced changes in flower and pod number and flower and fruit abscission in soybean[J]. Agron J, 1993, 85(2):221-225.
doi: 10.2134/agronj1993.00021962008500020011x URL |
| [2] |
Richards RA. Selectable traits to increase crop photosynjournal and yield of grain crops[J]. J Exp Bot, 2000, 51 Spec No:447-458.
doi: 10.1093/jexbot/51.suppl_1.447 URL |
| [3] |
Zhao DL, Atlin GN, Bastiaans L, et al. Developing selection protocols for weed competitiveness in aerobic rice[J]. Field Crops Res, 2006, 97(2/3):272-285.
doi: 10.1016/j.fcr.2005.10.008 URL |
| [4] |
Simon S, Morel K, Durand E, et al. Aphids at crossroads:when branch architecture alters aphid infestation patterns in the apple tree[J]. Trees, 2012, 26(1):273-282.
doi: 10.1007/s00468-011-0629-8 URL |
| [5] |
Zhao WG, Chao HB, Zhang LN, et al. Integration of QTL mapping and gene fishing techniques to dissect the multi-main stem trait in rapeseed(Brassica napus L.)[J]. Front Plant Sci, 2019, 10:1152.
doi: 10.3389/fpls.2019.01152 URL |
| [6] | 漆丽萍. 甘蓝型油菜株型与角果相关性状的QTL分析[D]. 武汉:华中农业大学, 2014. |
| Qi LP. Qtl analysis for the traits associated with plant architecture and silique in Brassica napus L.[D]. Wuhan:Huazhong Agricultural University, 2014. | |
| [7] |
Wang YH, Li JY. Genes controlling plant architecture[J]. Curr Opin Biotechnol, 2006, 17(2):123-129.
doi: 10.1016/j.copbio.2006.02.004 URL |
| [8] |
Doebley J, Stec A, Gustus C. Teosinte branched1 and the origin of maize:evidence for epistasis and the evolution of dominance[J]. Genetics, 1995, 141(1):333-346.
doi: 10.1093/genetics/141.1.333 pmid: 8536981 |
| [9] |
Hubbard L, McSteen P, Doebley J , et al. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte[J]. Genetics, 2002, 162(4):1927-1935.
doi: 10.1093/genetics/162.4.1927 pmid: 12524360 |
| [10] |
Aguilar-Martínez JA, Poza-Carrión C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds[J]. Plant Cell, 2007, 19(2):458-472.
pmid: 17307924 |
| [11] |
Dun EA, Brewer PB, Beveridge CA. Strigolactones:discovery of the elusive shoot branching hormone[J]. Trends Plant Sci, 2009, 14(7):364-372.
doi: 10.1016/j.tplants.2009.04.003 URL |
| [12] |
Leyser O. The control of shoot branching:an example of plant information processing[J]. Plant Cell Environ, 2009, 32(6):694-703.
doi: 10.1111/j.1365-3040.2009.01930.x URL |
| [13] |
Beveridge CA, Kyozuka J. New genes in the strigolactone-related shoot branching pathway[J]. Curr Opin Plant Biol, 2010, 13(1):34-39.
doi: 10.1016/j.pbi.2009.10.003 URL |
| [14] | Rameau C, Bertheloot J, Leduc N, et al. Multiple pathways regulate shoot branching[J]. Front Plant Sci, 2015, 5:741. |
| [15] |
Poza-Carrión C, Aguilar-Martínez JA, Cubas P. Role of TCP gene BRANCHED1 in the control of shoot branching in Arabidopsis[J]. Plant Signal Behav, 2007, 2(6):551-552.
doi: 10.4161/psb.2.6.4811 pmid: 19704556 |
| [16] |
Wang M, le Moigne MA, Bertheloot J, et al. BRANCHED1:a key hub of shoot branching[J]. Front Plant Sci, 2019, 10:76.
doi: 10.3389/fpls.2019.00076 URL |
| [17] |
Ma XL, Zhang QY, Zhu QL, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants[J]. Mol Plant, 2015, 8(8):1274-1284.
doi: 10.1016/j.molp.2015.04.007 URL |
| [18] | 杨阳. 油菜多室基因的鉴定及多室性状形成的分子调控机理解析[D]. 武汉:华中农业大学, 2019. |
| Yang Y. Identification of multilocular gene in Brassica napus and analysis of molecular regulation mechanism of multilocular traits[J]. Wuhan: Huazhong Agricultural University, 2019. | |
| [19] |
Liu Q, Wang C, Jiao XZ, et al. Hi-TOM:a platform for high-throu-ghput tracking of mutations induced by CRISPR/Cas systems[J]. Sci China Life Sci, 2019, 62(1):1-7.
doi: 10.1007/s11427-018-9402-9 URL |
| [20] |
Russell WL, Russell LB, Kelly EM. Radiation dose rate and mutation frequency[J]. Science, 1958, 128(3338):1546-1550.
doi: 10.1126/science.128.3338.1546 URL |
| [21] |
Sega GA. A review of the genetic effects of ethyl methanesulfonate[J]. Mutat Res, 1984, 134(2/3):113-142.
doi: 10.1016/0165-1110(84)90007-1 URL |
| [22] |
Yang Y, Zhu KY, Li HL, et al. Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development[J]. Plant Biotechnol J, 2018, 16(7):1322-1335.
doi: 10.1111/pbi.12872 pmid: 29250878 |
| [23] |
Wang YP, Cheng X, Shan QW, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew[J]. Nat Biotechnol, 2014, 32(9):947-951.
doi: 10.1038/nbt.2969 URL |
| [24] | Koonin EV, Makarova KS. Origins and evolution of CRISPR-Cas systems[J]. Philos Trans Royal Soc Lond Ser B Biol Sci, 2019, 374(1772):20180087. |
| [25] |
Braun N, de Saint Germain A, Pillot JP, et al. The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching[J]. Plant Physiol, 2012, 158(1):225-238.
doi: 10.1104/pp.111.182725 pmid: 22045922 |
| [26] |
Dun EA, de Saint Germain A, Rameau C, et al. Antagonistic action of strigolactone and cytokinin in bud outgrowth control[J]. Plant Physiol, 2012, 158(1):487-498.
doi: 10.1104/pp.111.186783 URL |
| [27] |
Lantzouni O, Klermund C, Schwechheimer C. Largely additive effects of gibberellin and strigolactone on gene expression in Arabidopsis thaliana seedlings[J]. Plant J, 2017, 92(5):924-938.
doi: 10.1111/tpj.13729 URL |
| [1] | WANG Bao-bao, WANG Hai-yang. Molecular Design of Ideal Plant Architecture for High-density Tolerance of Maize Plant [J]. Biotechnology Bulletin, 2023, 39(8): 11-30. |
| [2] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [3] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [4] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [5] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [6] | CAI Meng-xian, GAO Zuo-min, HU Li-juan, FENG Qun, WANG Hong-cheng, ZHU Bin. Development and Genetic Analysis of Two Nullisomic Lines(NC1 and NC2)in Natural Brassica napus [J]. Biotechnology Bulletin, 2023, 39(3): 81-88. |
| [7] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [8] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [9] | ZHI Tian-tian, ZHOU Zhou, CHEN Ji-peng, HAN Cheng-yun. Cloning, Functional Identification and Expression Analysis of FAH, a Key Gene for Tyrosine Metabolism in Brassica napus L. [J]. Biotechnology Bulletin, 2023, 39(10): 115-127. |
| [10] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [11] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [12] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [13] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [14] | LIN Ke-yun, DUAN Yu-jing, WANG Gao-sheng, SUN Nian-li, FANG Yu-jie, WANG You-ping. Cloning and Functional Identification of BnNF-YA1 in Brassica napus L. [J]. Biotechnology Bulletin, 2022, 38(4): 106-116. |
| [15] | JIN Jiao-jiao, LIU Zi-gang, MI Wen-bo, XU Ming-xia, ZOU Ya, XU Chun-mei, ZHAO Cai-xia. Identification of Low Temperature Stress-responsive Genes Regulating Photosynthetic Characteristics in the Leaves of Brassica napus by RNA-Seq [J]. Biotechnology Bulletin, 2022, 38(4): 126-142. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||