








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (7): 1-12.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0071
XUE Man-de1,2( ), ZHAO Feng-yue1,2, LI Jie1,2, JIANG Dan-hua1,2(
), ZHAO Feng-yue1,2, LI Jie1,2, JIANG Dan-hua1,2( )
)
Received:2022-01-17
Online:2022-07-26
Published:2022-08-09
Contact:
JIANG Dan-hua
E-mail:xuemande@genetics.ac.cn;dhjiang@genetics.ac.cn
XUE Man-de, ZHAO Feng-yue, LI Jie, JIANG Dan-hua. Advances in Histone Variants in Plant Epigenetic Regulation[J]. Biotechnology Bulletin, 2022, 38(7): 1-12.
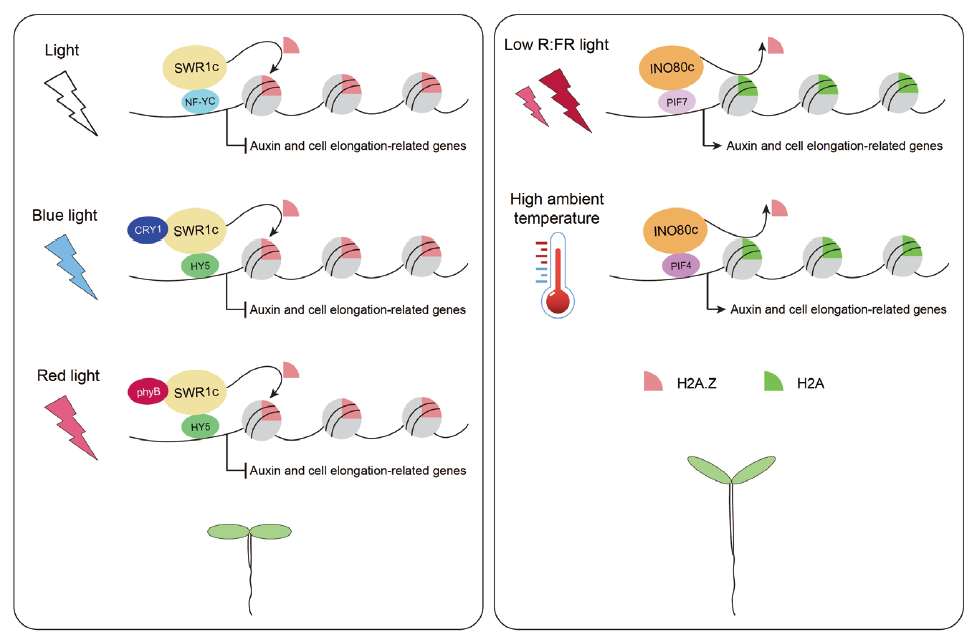
Fig. 2 Role of H2A.Z in photomorphogenesis,shade avo-idance and thermomorphogenesis of A. thaliana Under light conditions,the SWR1 complex interacts with transcription factors and photoreceptors,and deposits H2A.Z at auxin and cell elongation-related genes to repress their expression,resulting in photomorphogenesis. Under low R:FR light or high ambient temperature condition,the INO80 complex interacts with PIF proteins and mediates H2A.Z eviction and transcription activation at their targets,leading to shade avoidance response or thermomorphogenesis respectively
| [1] |
Borg M, Jiang DH, Berger F. Histone variants take center stage in shaping the epigenome[J]. Curr Opin Plant Biol, 2021, 61:101991.
doi: 10.1016/j.pbi.2020.101991 URL |
| [2] |
Jiang DH, Berger F. Histone variants in plant transcriptional regulation[J]. Biochim Biophys Acta Gene Regul Mech, 2017, 1860(1):123-130.
doi: 10.1016/j.bbagrm.2016.07.002 URL |
| [3] |
Kawashima T, Lorković ZJ, Nishihama R, et al. Diversification of histone H2A variants during plant evolution[J]. Trends Plant Sci, 2015, 20(7):419-425.
doi: 10.1016/j.tplants.2015.04.005 pmid: 25983206 |
| [4] | Coleman-Derr D, Zilberman D. Deposition of histone variant H2A. Z within gene bodies regulates responsive genes[J]. PLoS Genet, 2012, 8(10):e1002988. |
| [5] |
Dai XZ, Bai YH, Zhao LH, et al. H2A. Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in Arabidopsis[J]. Mol Plant, 2017, 10(10):1274-1292.
doi: 10.1016/j.molp.2017.09.007 URL |
| [6] | Giaimo BD, Ferrante F, Herchenröther A, et al. The histone variant H2A. Z in gene regulation[J]. Epigenetics Chromatin, 2019,(1):37. |
| [7] |
Hu GQ, Cui KR, Northrup D, et al. H2A. Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation[J]. Cell Stem Cell, 2013, 12(2):180-192.
doi: 10.1016/j.stem.2012.11.003 URL |
| [8] |
Raisner RM, Hartley PD, Meneghini MD, et al. Histone variant H2A. Z marks the 5' ends of both active and inactive genes in euchromatin[J]. Cell, 2005, 123(2):233-248.
pmid: 16239142 |
| [9] |
Gómez-Zambrano Á, Merini W, Calonje M. The repressive role of Arabidopsis H2A. Z in transcriptional regulation depends on AtBMI1 activity[J]. Nat Commun, 2019, 10(1):2828.
doi: 10.1038/s41467-019-10773-1 pmid: 31249301 |
| [10] |
Choi K, Park C, Lee J, et al. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development[J]. Development, 2007, 134(10):1931-1941.
doi: 10.1242/dev.001891 URL |
| [11] |
Gómez-Zambrano Á, Crevillén P, Franco-Zorrilla JM, et al. Arabidopsis SWC4 binds DNA and recruits the SWR1 complex to modulate histone H2A. Z deposition at key regulatory genes[J]. Mol Plant, 2018, 11(6):815-832.
doi: S1674-2052(18)30122-9 pmid: 29604400 |
| [12] |
Nie WF, Lei MG, Zhang MX, et al. Histone acetylation recruits the SWR1 complex to regulate active DNA demethylation in Arabidopsis[J]. Proc Natl Acad Sci USA, 2019, 116(33):16641-16650.
doi: 10.1073/pnas.1906023116 URL |
| [13] | Sijacic P, Holder DH, Bajic M, et al. Methyl-CpG-binding domain 9(MBD9)is required for H2A. Z incorporation into chromatin at a subset of H2A. Z-enriched regions in the Arabidopsisgenome[J]. PLoS Genet, 2019, 15(8):e1008326. |
| [14] | Luo YX, Hou XM, Zhang CJ, et al. A plant-specific SWR1 chromatin-remodeling complex couples histone H2A. Z deposition with nucleosome sliding[J]. EMBO J, 2020, 39(7):e102008. |
| [15] |
Papamichos-Chronakis M, Watanabe S, Rando OJ, et al. Global regulation of H2A. Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity[J]. Cell, 2011, 144(2):200-213.
doi: 10.1016/j.cell.2010.12.021 pmid: 21241891 |
| [16] | Tramantano M, Sun L, Au C, et al. Constitutive turnover of histone H2A. Z at yeast promoters requires the preinitiation complex[J]. eLife, 2016, 5:e14243. |
| [17] | Ranjan A, Nguyen VQ, Liu S, et al. Live-cell single particle imaging reveals the role of RNA polymerase II in histone H2A. Z eviction[J]. eLife, 2020, 9:e55667. |
| [18] |
Zhang C, Cao L, Rong L, et al. The chromatin-remodeling factor AtINO80 plays crucial roles in genome stability maintenance and in plant development[J]. Plant J, 2015, 82(4):655-668.
doi: 10.1111/tpj.12840 URL |
| [19] |
Wang J, Gao S, Peng X, et al. Roles of the INO80 and SWR1 chromatin remodeling complexes in plants[J]. Int J Mol Sci, 2019, 20(18):4591.
doi: 10.3390/ijms20184591 URL |
| [20] |
Xue MD, Zhang HR, Zhao FY, et al. The INO80 chromatin remodeling complex promotes thermomorphogenesis by connecting H2A. Z eviction and active transcription in Arabidopsis[J]. Mol Plant, 2021, 14(11):1799-1813.
doi: 10.1016/j.molp.2021.07.001 URL |
| [21] |
Yang CW, Yin LF, Xie FM, et al. AtINO80 represses photomorphogenesis by modulating nucleosome density and H2A. Z incorporation in light-related genes[J]. Proc Natl Acad Sci USA, 2020, 117(52):33679-33688.
doi: 10.1073/pnas.2001976117 URL |
| [22] |
Wang YF, Zhong ZH, Zhang YX, et al. NAP1-RELATED PROTEIN1 and 2 negatively regulate H2A. Z abundance in chromatin in Arabidopsis[J]. Nat Commun, 2020, 11(1):2887.
doi: 10.1038/s41467-020-16691-x URL |
| [23] |
Du KX, Luo Q, Yin LF, et al. OsChz1 acts as a histone chaperone in modulating chromatin organization and genome function in rice[J]. Nat Commun, 2020, 11(1):5717.
doi: 10.1038/s41467-020-19586-z URL |
| [24] |
Li C, Liu YH, Shen WH, et al. Chromatin-remodeling factor OsINO80 is involved in regulation of gibberellin biosynthesis and is crucial for rice plant growth and development[J]. J Integr Plant Biol, 2018, 60(2):144-159.
doi: 10.1111/jipb.12603 URL |
| [25] |
Mao Z, Pan L, Wang WX, et al. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A. Z[J]. Cell Res, 2014, 24(4):389-399.
doi: 10.1038/cr.2014.30 URL |
| [26] |
Obri A, Ouararhni K, Papin C, et al. ANP32E is a histone chaperone that removes H2A. Z from chromatin[J]. Nature, 2014, 505(7485):648-653.
doi: 10.1038/nature12922 URL |
| [27] |
Deal RB, Topp CN, McKinney EC, et al. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A. Z[J]. Plant Cell, 2007, 19(1):74-83.
doi: 10.1105/tpc.106.048447 URL |
| [28] |
Choi K, Kim J, Hwang HJ, et al. The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors[J]. Plant Cell, 2011, 23(1):289-303.
doi: 10.1105/tpc.110.075911 URL |
| [29] | Xu ML, Leichty AR, Hu TQ, et al. H2A. Z promotes the transcription of MIR156A and MIR156C in Arabidopsis by facilitating the deposition of H3K4me3[J]. Development, 2018, 145(2):dev152868. |
| [30] | Zander M, Willige BC, He YP, et al. Epigenetic silencing of a multifunctional plant stress regulator[J]. eLife, 2019, 8:e47835. |
| [31] |
Cai HY, Zhang M, Chai MN, et al. Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A. Z and H3K4me3[J]. New Phytol, 2019, 221(1):295-308.
doi: 10.1111/nph.15306 URL |
| [32] |
Yang XD, Zhang XL, Yang YX, et al. The histone variant Sl_H2A. Z regulates carotenoid biosynthesis and gene expression during tomato fruit ripening[J]. Hortic Res, 2021, 8(1):85.
doi: 10.1038/s41438-021-00520-3 URL |
| [33] |
March-Díaz R, García-Domínguez M, Lozano-Juste J, et al. Histone H2A. Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis[J]. Plant J, 2008, 53(3):475-487.
pmid: 17988222 |
| [34] |
Cai HY, Huang YM, Chen FQ, et al. ERECTA signaling regulates plant immune responses via chromatin-mediated promotion of WRKY33 binding to target genes[J]. New Phytol, 2021, 230(2):737-756.
doi: 10.1111/nph.17200 URL |
| [35] |
Sura W, Kabza M, Karlowski WM, et al. Dual role of the histone variant H2A. Z in transcriptional regulation of stress-response genes[J]. Plant Cell, 2017, 29(4):791-807.
doi: 10.1105/tpc.16.00573 URL |
| [36] |
Nguyen NH, Cheong JJ. H2A. Z-containing nucleosomes are evicted to activate AtMYB44 transcription in response to salt stress[J]. Biochem Biophys Res Commun, 2018, 499(4):1039-1043.
doi: 10.1016/j.bbrc.2018.04.048 URL |
| [37] |
Zhu ZQ, Lin CT. Photomorphogenesis:When blue meets red[J]. Nat Plants, 2016, 2:16019.
doi: 10.1038/nplants.2016.19 URL |
| [38] |
Zhang CY, Qian Q, Huang X, et al. NF-YCs modulate histone variant H2A. Z deposition to regulate photomorphogenic growth in Arabidopsis[J]. J Integr Plant Biol, 2021, 63(6):1120-1132.
doi: 10.1111/jipb.13109 URL |
| [39] | Nemhauser J, Chory J. Photomorphogenesis[J]. Arabidopsis Book, 2002, 1:e0054. |
| [40] |
Mao ZL, Wei XX, Li L, et al. Arabidopsis cryptochrome 1 controls photomorphogenesis through regulation of H2A. Z deposition[J]. Plant Cell, 2021, 33(6):1961-1979.
doi: 10.1093/plcell/koab091 URL |
| [41] |
Wei XX, Wang WT, Xu P, et al. Phytochrome B interacts with SWC6 and ARP6 to regulate H2A. Z deposition and photomorphogensis in Arabidopsis[J]. J Integr Plant Biol, 2021, 63(6):1133-1146.
doi: 10.1111/jipb.13111 URL |
| [42] |
Willige BC, Zander M, Yoo CY, et al. PHYTOCHROME-INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants[J]. Nat Genet, 2021, 53(7):955-961.
doi: 10.1038/s41588-021-00882-3 pmid: 34140685 |
| [43] |
Li X, Liang T, Liu HT. How plants coordinate their development in response to light and temperature signals[J]. Plant Cell, 2022, 34(3):955-966.
doi: 10.1093/plcell/koab302 URL |
| [44] |
Kumar SV, Wigge PA. H2A. Z-containing nucleosomes mediate the thermosensory response in Arabidopsis[J]. Cell, 2010, 140(1):136-147.
doi: 10.1016/j.cell.2009.11.006 URL |
| [45] |
Shang JY, Lu YJ, Cai XW, et al. COMPASS functions as a module of the INO80 chromatin remodeling complex to mediate histone H3K4 methylation in Arabidopsis[J]. Plant Cell, 2021, 33(10):3250-3271.
doi: 10.1093/plcell/koab187 URL |
| [46] |
Zahraeifard S, Foroozani M, Sepehri A, et al. Rice H2A. Z negatively regulates genes responsive to nutrient starvation but promotes expression of key housekeeping genes[J]. J Exp Bot, 2018, 69(20):4907-4919.
doi: 10.1093/jxb/ery244 pmid: 29955860 |
| [47] | Boden SA, Kavanová M, Finnegan EJ, et al. Thermal stress effects on grain yield in Brachypodium distachyon occur via H2A. Z-nucleosomes[J]. Genome Biol, 2013, 14(6):R65. |
| [48] |
del Olmo I, Poza-Viejo L, Piñeiro M, et al. High ambient temperature leads to reduced FT expression and delayed flowering in Brassica rapa via a mechanism associated with H2A. Z dynamics[J]. Plant J, 2019, 100(2):343-356.
doi: 10.1111/tpj.14446 URL |
| [49] | 杨学东, 田守波, 朱为民, 等. 番茄组蛋白变体H2A. Z基因双突变体的获得及其功能研究[J]. 园艺学报, 2018, 45(6):1081-1088. |
| Yang XD, Tian SB, Zhu WM, et al. Tomato double mutant of histone variant gene was obtained and its function research[J]. Acta Hortic Sin, 2018, 45(6):1081-1088. | |
| [50] |
Bewersdorf J, Bennett BT, Knight KL. H2AX chromatin structures and their response to DNA damage revealed by 4Pi microscopy[J]. Proc Natl Acad Sci USA, 2006, 103(48):18137-18142.
doi: 10.1073/pnas.0608709103 URL |
| [51] |
Yelagandula R, Stroud H, Holec S, et al. The histone variant H2A. W defines heterochromatin and promotes chromatin condensation in Arabidopsis[J]. Cell, 2014, 158(1):98-109.
doi: 10.1016/j.cell.2014.06.006 pmid: 24995981 |
| [52] |
Lang J, Smetana O, Sanchez-Calderon L, et al. Plant γH2AX foci are required for proper DNA DSB repair responses and colocalize with E2F factors[J]. New Phytol, 2012, 194(2):353-363.
doi: 10.1111/j.1469-8137.2012.04062.x URL |
| [53] |
Piquet S le Parc F, Bai SK, et al. The histone chaperone FACT coordinates H2A. X-dependent signaling and repair of DNA damage[J]. Mol Cell, 2018, 72(5):888-901. e7.
doi: 10.1016/j.molcel.2018.09.010 URL |
| [54] |
Bourguet P, Picard CL, Yelagandula R, et al. The histone variant H2A. W and linker histone H1 co-regulate heterochromatin accessibility and DNA methylation[J]. Nat Commun, 2021, 12(1):2683.
doi: 10.1038/s41467-021-22993-5 pmid: 33976212 |
| [55] |
Osakabe A, Jamge B, Axelsson E, et al. The chromatin remodeler DDM1 prevents transposon mobility through deposition of histone variant H2A. W[J]. Nat Cell Biol, 2021, 23(4):391-400.
doi: 10.1038/s41556-021-00658-1 URL |
| [56] |
Lorković ZJ, Park C, Goiser M, et al. Compartmentalization of DNA damage response between heterochromatin and euchromatin is mediated by distinct H2A histone variants[J]. Curr Biol, 2017, 27(8):1192-1199.
doi: S0960-9822(17)30271-3 pmid: 28392109 |
| [57] |
Dalal Y, Bui M. Down the rabbit hole of centromere assembly and dynamics[J]. Curr Opin Cell Biol, 2010, 22(3):392-402.
doi: 10.1016/j.ceb.2010.02.005 URL |
| [58] |
Lermontova I, Schubert V, Fuchs J, et al. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain[J]. Plant Cell, 2006, 18(10):2443-2451.
pmid: 17028205 |
| [59] |
Fukagawa T, Earnshaw WC. The centromere:chromatin foundation for the kinetochore machinery[J]. Dev Cell, 2014, 30(5):496-508.
doi: 10.1016/j.devcel.2014.08.016 pmid: 25203206 |
| [60] |
Lermontova I, Koroleva O, Rutten T, et al. Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation[J]. Plant J, 2011, 68(1):40-50.
doi: 10.1111/j.1365-313X.2011.04664.x URL |
| [61] |
Teo CH, Lermontova I, Houben A, et al. De novo generation of plant centromeres at tandem repeats[J]. Chromosoma, 2013, 122(3):233-241.
doi: 10.1007/s00412-013-0406-0 URL |
| [62] |
Ravi M, Chan SWL. Haploid plants produced by centromere-mediated genome elimination[J]. Nature, 2010, 464(7288):615-618.
doi: 10.1038/nature08842 URL |
| [63] |
Lv J, Yu K, Wei J, et al. Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3[J]. Nat Biotechnol, 2020, 38(12):1397-1401.
doi: 10.1038/s41587-020-0728-4 URL |
| [64] | Wang N, Gent JI, Dawe RK. Haploid induction by a maize cenh3 null mutant[J]. Sci Adv, 2021, 7(4):eabe2299. |
| [65] |
Karimi-Ashtiyani R, Ishii T, Niessen M, et al. Point mutation impairs centromeric CENH3 loading and induces haploid plants[J]. Proc Natl Acad Sci USA, 2015, 112(36):11211-11216.
doi: 10.1073/pnas.1504333112 URL |
| [66] | Kuppu S, Tan EH, Nguyen H, et al. Point mutations in centromeric histone induce post-zygotic incompatibility and uniparental inheritance[J]. PLoS Genet, 2015, 11(9):e1005494. |
| [67] | Maheshwari S, Tan EH, West A, et al. Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids[J]. PLoS Genet, 2015, 11(1):e1004970. |
| [68] |
Talbert PB, Ahmad K, Almouzni G, et al. A unified phylogeny-based nomenclature for histone variants[J]. Epigenetics Chromatin, 2012, 5:7.
doi: 10.1186/1756-8935-5-7 pmid: 22650316 |
| [69] |
Lu L, Chen XS, Qian SM, et al. The plant-specific histone residue Phe41 is important for genome-wide H3. 1 distribution[J]. Nat Commun, 2018, 9(1):630.
doi: 10.1038/s41467-017-02454-8 URL |
| [70] | Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3. 3[J]. Cell Res, 2011, 21(3):421-434. |
| [71] |
Tagami H, Ray-Gallet D, Almouzni G, et al. Histone H3. 1 and H3. 3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis[J]. Cell, 2004, 116(1):51-61.
doi: 10.1016/S0092-8674(03)01064-X URL |
| [72] |
Ahmad K, Henikoff S. The histone variant H3. 3 marks active chromatin by replication-independent nucleosome assembly[J]. Mol Cell, 2002, 9(6):1191-1200.
pmid: 12086617 |
| [73] |
Nie X, Wang HF, Li J, et al. The HIRA complex that deposits the histone H3. 3 is conserved in Arabidopsis and facilitates transcriptional dynamics[J]. Biol Open, 2014, 3(9):794-802.
doi: 10.1242/bio.20148680 URL |
| [74] |
Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin[J]. Cell, 1999, 96(4):575-585.
doi: 10.1016/s0092-8674(00)80661-3 pmid: 10052459 |
| [75] |
Duc C, Benoit M, Détourné G, et al. Arabidopsis ATRX modulates H3. 3 occupancy and fine-tunes gene expression[J]. Plant Cell, 2017, 29(7):1773-1793.
doi: 10.1105/tpc.16.00877 URL |
| [76] |
Lewis PW, Elsaesser SJ, Noh KM, et al. Daxx is an H3. 3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres[J]. Proc Natl Acad Sci USA, 2010, 107(32):14075-14080.
doi: 10.1073/pnas.1008850107 URL |
| [77] |
Wang HF, Jiang DH, Axelsson E, et al. LHP1 interacts with ATRX through plant-specific domains at specific loci targeted by PRC2[J]. Mol Plant, 2018, 11(8):1038-1052.
doi: 10.1016/j.molp.2018.05.004 URL |
| [78] |
Stroud H, Otero S, Desvoyes B, et al. Genome-wide analysis of histone H3. 1 and H3. 3 variants in Arabidopsis thaliana[J]. Proc Natl Acad Sci USA, 2012, 109(14):5370-5375.
doi: 10.1073/pnas.1203145109 URL |
| [79] | Wollmann H, Holec S, Alden K, et al. Dynamic deposition of histone variant H3. 3 accompanies developmental remodeling of the Arabidopsis transcriptome[J]. PLoS Genet, 2012, 8(5):e1002658. |
| [80] |
Shu H, Nakamura M, Siretskiy A, et al. Arabidopsis replacement histone variant H3. 3 occupies promoters of regulated genes[J]. Genome Biol, 2014, 15(4):R62.
doi: 10.1186/gb-2014-15-4-r62 URL |
| [81] |
Mendiratta S, Gatto A, Almouzni G. Histone supply:Multitiered regulation ensures chromatin dynamics throughout the cell cycle[J]. J Cell Biol, 2019, 218(1):39-54.
doi: 10.1083/jcb.201807179 pmid: 30257851 |
| [82] |
Reverón-Gómez N, González-Aguilera C, Stewart-Morgan KR, et al. Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication[J]. Mol Cell, 2018, 72(2):239-249. e5.
doi: S1097-2765(18)30644-0 pmid: 30146316 |
| [83] |
Jacob Y, Feng SH, LeBlanc CA, et al. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing[J]. Nat Struct Mol Biol, 2009, 16(7):763-768.
doi: 10.1038/nsmb.1611 URL |
| [84] |
Jacob Y, Bergamin E, Donoghue MTA, et al. Selective methylation of histone H3 variant H3. 1 regulates heterochromatin replication[J]. Science, 2014, 343(6176):1249-1253.
doi: 10.1126/science.1248357 URL |
| [85] |
Jiang DH, Berger F. DNA replication-coupled histone modification maintains Polycomb gene silencing in plants[J]. Science, 2017, 357(6356):1146-1149.
doi: 10.1126/science.aan4965 URL |
| [86] |
Raynaud C, Sozzani R, Glab N, et al. Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen(PCNA)in Arabidopsis[J]. Plant J, 2006, 47(3):395-407.
pmid: 16771839 |
| [87] |
Dong J, LeBlanc C, Poulet A, et al. H3. 1K27me1 maintains transcriptional silencing and genome stability by preventing GCN5-mediated histone acetylation[J]. Plant Cell, 2021, 33(4):961-979.
doi: 10.1093/plcell/koaa027 URL |
| [88] |
Banaszynski LA, Wen DC, Dewell S, et al. Hira-dependent histone H3. 3 deposition facilitates PRC2 recruitment at developmental loci in ES cells[J]. Cell, 2013, 155(1):107-120.
doi: 10.1016/j.cell.2013.08.061 pmid: 24074864 |
| [89] |
Sitbon D, Boyarchuk E, Dingli F, et al. Histone variant H3. 3 residue S31 is essential for Xenopus gastrulation regardless of the deposition pathway[J]. Nat Commun, 2020, 11(1):1256.
doi: 10.1038/s41467-020-15084-4 pmid: 32152320 |
| [90] |
Otero S, Desvoyes B, Peiró R, et al. Histone H3 dynamics reveal domains with distinct proliferation potential in the Arabidopsis root[J]. Plant Cell, 2016, 28(6):1361-1371.
doi: 10.1105/tpc.15.01003 URL |
| [91] |
Lee LR, Wengier DL, Bergmann DC. Cell-type-specific transcriptome and histone modification dynamics during cellular reprogramming in the Arabidopsis stomatal lineage[J]. Proc Natl Acad Sci USA, 2019, 116(43):21914-21924.
doi: 10.1073/pnas.1911400116 URL |
| [92] |
Wollmann H, Stroud H, Yelagandula R, et al. The histone H3 variant H3. 3 regulates gene body DNA methylation in Arabidopsis thaliana[J]. Genome Biol, 2017, 18(1):94.
doi: 10.1186/s13059-017-1221-3 pmid: 28521766 |
| [93] |
Zhao FY, Zhang HR, Zhao T, et al. The histone variant H3. 3 promotes the active chromatin state to repress flowering in Arabidopsis[J]. Plant Physiol, 2021, 186(4):2051-2063.
doi: 10.1093/plphys/kiab224 URL |
| [94] | Yan A, Borg M, Berger F, et al. The atypical histone variant H3. 15 promotes callus formation in Arabidopsis thaliana[J]. Development, 2020, 147(11):dev184895. |
| [95] |
Borg M, Jacob Y, Susaki D, et al. Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin[J]. Nat Cell Biol, 2020, 22(6):621-629.
doi: 10.1038/s41556-020-0515-y URL |
| [96] |
Ingouff M, Rademacher S, Holec S, et al. Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis[J]. Curr Biol, 2010, 20(23):2137-2143.
doi: 10.1016/j.cub.2010.11.012 URL |
| [97] |
Ingouff M, Hamamura Y, Gourgues M, et al. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants[J]. Curr Biol, 2007, 17(12):1032-1037.
doi: 10.1016/j.cub.2007.05.019 URL |
| [98] |
Zhao P, Zhou XM, Shen K, et al. Two-step maternal-to-zygotic transition with two-phase parental genome contributions[J]. Dev Cell, 2019, 49(6):882-893. e5.
doi: 10.1016/j.devcel.2019.04.016 URL |
| [99] | Jiang DH, Borg M, Lorković ZJ, et al. The evolution and functional divergence of the histone H2B family in plants[J]. PLoS Genet, 2020, 16(7):e1008964. |
| [100] | Khadka J, Pesok A, Grafi G. Plant histone HTB(H2B)variants in regulating chromatin structure and function[J]. Plants(Basel), 2020, 2 9(11):1435. |
| [101] |
Zhou BR, Jiang JS, Feng HQ, et al. Structural mechanisms of nucleosome recognition by linker histones[J]. Mol Cell, 2015, 59(4):628-638.
doi: 10.1016/j.molcel.2015.06.025 URL |
| [102] |
Bednar J, Garcia-Saez I, Boopathi R, et al. Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1[J]. Mol Cell, 2017, 66(5):729.
doi: S1097-2765(17)30358-1 pmid: 28575663 |
| [103] |
Kotliński M, Knizewski L, Muszewska A, et al. Phylogeny-based systematization of Arabidopsis proteins with histone H1 globular domain[J]. Plant Physiol, 2017, 174(1):27-34.
doi: 10.1104/pp.16.00214 pmid: 28298478 |
| [104] |
Rutowicz K, Puzio M, Halibart-Puzio J, et al. A specialized histone H1 variant is required for adaptive responses to complex abiotic stress and related DNA methylation in Arabidopsis[J]. Plant Physiol, 2015, 169(3):2080-2101.
doi: 10.1104/pp.15.00493 pmid: 26351307 |
| [105] |
Zemach A, Kim MY, Hsieh PH, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin[J]. Cell, 2013, 153(1):193-205.
doi: 10.1016/j.cell.2013.02.033 pmid: 23540698 |
| [106] |
Rutowicz K, Lirski M, Mermaz B, et al. Linker histones are fine-scale chromatin architects modulating developmental decisions in Arabidopsis[J]. Genome Biol, 2019, 20(1):157.
doi: 10.1186/s13059-019-1767-3 pmid: 31391082 |
| [107] |
Zhang PX, Li XL, Wang YF, et al. PRMT6 physically associates with nuclear factor Y to regulate photoperiodic flowering in Arabidopsis[J]. aBIOTECH, 2021, 2(4):403-414.
doi: 10.1007/s42994-021-00065-y URL |
| [108] | Zhao T, Zhan ZP, Jiang DH. Histone modifications and their regulatory roles in plant development and environmental memory[J]. J Genet Genomics, 2019, 46(10):467-476. |
| [1] | ZHAN Yan, ZHOU Li-bin, JIN Wen-jie, DU Yan, YU Li-xia, QU Ying, MA Yong-gui, LIU Rui-yuan. Research Progress in Plant Leaf Color Mutation Induced by Radiation [J]. Biotechnology Bulletin, 2023, 39(8): 106-113. |
| [2] | WANG Bao-bao, WANG Hai-yang. Molecular Design of Ideal Plant Architecture for High-density Tolerance of Maize Plant [J]. Biotechnology Bulletin, 2023, 39(8): 11-30. |
| [3] | JIANG Run-hai, JIANG Ran-ran, ZHU Cheng-qiang, HOU Xiu-li. Research Progress in Mechanisms of Microbial-enhanced Phytoremediation for Lead-contaminated Soil [J]. Biotechnology Bulletin, 2023, 39(8): 114-125. |
| [4] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [5] | LIU Bao-cai, CHEN Jing-ying, ZHANG Wu-jun, HUANG Ying-zhen, ZHAO Yun-qing, LIU Jian-chao, WEI Zhi-cheng. Characteristics Analysis of Seed Microrhizome Gene Expression of Polygonatum cyrtonema [J]. Biotechnology Bulletin, 2023, 39(8): 220-233. |
| [6] | SHI Jia-xin, LIU Kai, ZHU Jin-jie, QI Xian-tao, XIE Chuan-xiao, LIU Chang-lin. Gene Editing Reshaping Maize Plant Type for Increasing Hybrid Yield [J]. Biotechnology Bulletin, 2023, 39(8): 62-69. |
| [7] | ZHANG Yong, XU Tian-jun, LYU Tian-fang, XING Jin-feng, LIU Hong-wei, CAI Wan-tao, LIU Yue-e, ZHAO Jiu-ran, WANG Rong-huan. Effects of Planting Density on the Stem Quality and Root Phenotypic Characters of Summer Sowing Maize [J]. Biotechnology Bulletin, 2023, 39(8): 70-79. |
| [8] | YAO Sha-sha, WANG Jing-jing, WANG Jun-jie, LIANG Wei-hong. Molecular Mechanisms of Rice Grain Size Regulation Related to Plant Hormone Signaling Pathways [J]. Biotechnology Bulletin, 2023, 39(8): 80-90. |
| [9] | ZHANG Man, ZHANG Ye-zhuo, HE Qi-zou-hong, E Yi-lan, LI Ye. Advances in Plant Cell Wall Structure and Imaging Technology [J]. Biotechnology Bulletin, 2023, 39(7): 113-122. |
| [10] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [11] | XU Jian-xia, DING Yan-qing, FENG Zhou, CAO Ning, CHENG Bin, GAO Xu, ZOU Gui-hua, ZHANG Li-yi. QTL Mapping of Sorghum Plant Height and Internode Numbers Based on Super-GBS Technique [J]. Biotechnology Bulletin, 2023, 39(7): 185-194. |
| [12] | LI Yu-ling, MAO Xin, ZHANG Yuan-shuai, DONG Yuan-fu, LIU Cui-lan, DUAN Chun-hua, MAO Xiu-hong. Applications and Perspectives of Radiation Mutagenesis in Woody Plant Breeding [J]. Biotechnology Bulletin, 2023, 39(6): 12-30. |
| [13] | YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum [J]. Biotechnology Bulletin, 2023, 39(6): 259-273. |
| [14] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [15] | LUO Yi, ZHANG Li-juan, HUANG Wei, WANG Ning, Wuerlika MAITIHASEM, SHI Chong, WANG Wei. Identification of a Uranium-resistant Strain and Its Growth-promoting Properties [J]. Biotechnology Bulletin, 2023, 39(5): 286-296. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||