








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (9): 158-166.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1444
Previous Articles Next Articles
CHEN Guang1,2,3( ), LI Jia4, DU Rui-ying1,2,3, WANG Xu1,2,3(
), LI Jia4, DU Rui-ying1,2,3, WANG Xu1,2,3( )
)
Received:2021-11-18
Online:2022-09-26
Published:2022-10-11
Contact:
WANG Xu
E-mail:chenguang0066@126.com;wangxuguangzhou@126.com
CHEN Guang, LI Jia, DU Rui-ying, WANG Xu. Identification and Gene Functional Analysis of Salinity-hypersensitive Mutant ss2 in Rice[J]. Biotechnology Bulletin, 2022, 38(9): 158-166.
| 基因名称 Gene name | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| UBQ5 | CTCGCCGACTACAACATCCA | TCTTGGGCTTGGTGTACGTCTT |
| OsSUT4 | CGCCGGCGGTGGCGGCCTCA | CGTGAGGAGCGAGAGCTGA |
| OsSWEET11 | GACGTTCTTGCAGGTGTACA | TAGCGGACGATGTAGGCGGC |
| OsSWEET14 | TTCCCAACGTGCTGGGCTTCT | GCACCTCGCGGGTCTTGACG |
| OsMT | GCTGCCAGGCAGGAAGCT | GGTTCCAGTTTCACCACGACA |
Table 1 Primer sequences used for RT-qPCR assays
| 基因名称 Gene name | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| UBQ5 | CTCGCCGACTACAACATCCA | TCTTGGGCTTGGTGTACGTCTT |
| OsSUT4 | CGCCGGCGGTGGCGGCCTCA | CGTGAGGAGCGAGAGCTGA |
| OsSWEET11 | GACGTTCTTGCAGGTGTACA | TAGCGGACGATGTAGGCGGC |
| OsSWEET14 | TTCCCAACGTGCTGGGCTTCT | GCACCTCGCGGGTCTTGACG |
| OsMT | GCTGCCAGGCAGGAAGCT | GGTTCCAGTTTCACCACGACA |
| 标记 Marker | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| SS11 | GCAACTGGTGGAGTCTATTT | CATGCTAACATGAGGTGATC |
| SS12 | ACGCCTCCCAAGTCGAAAGG | GGTGGGCCTCGATTGTAAGTAG |
| SS10 | TTGGCTCTTCTCCTTAGTAT | CATTTGTATCTTGTGAACGT |
| SS3 | CCAATGTTTGCTCCAGAT | TTCAATGACCCACGTCCC |
| SS4 | TGGTTTTCCTTGTTGCTG | GCTTGCGGCTCTGCTTAC |
| SS20 | TACAGGTATGCTGCTTTTCC | CTGGTCCTTTTCATTCTAAC |
| SS21 | AAAACATGCTCCAACAGCCT | CCAAATGTAGCCAGTGAGGA |
| SS22 | GGAGGAGTTCATTTGAGGCG | CTGGGTGGGCTAGGAAGTAA |
| SS23 | ATACCTCCTTGTATTCGCACT | CGATCGATTGCCACATTATA |
| SS24 | AGCGTGAATCTAATAGCACT | CGTTCAACAAGACCCAATAC |
| SS25 | TTGTAACCGTCGATTTCGTTC | CCGCTCCGTCACTCTACTACC |
| SS16 | CCTCCGACCTCAGCACCTGC | GTTGGCGTCCGCTGCTCCTG |
| SS17 | AGGTAGGCGTGGCGATCAAC | CTTCTCCGGTCACCATCCAC |
| SS27 | TTGGCGATTAATGATCCGGGAAC | CGTTCGTGCCGGTGATGTCG |
| SS29 | ACAACAGTTCTTCACCAGAG | GTAGTATAAATTGTAATAGCTCAA |
| SS32 | GTTAAATGAATCATCAGGAT | AGTAGTCTTGAATTCGCTGT |
Table 2 Marked primer used for gene mapping
| 标记 Marker | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| SS11 | GCAACTGGTGGAGTCTATTT | CATGCTAACATGAGGTGATC |
| SS12 | ACGCCTCCCAAGTCGAAAGG | GGTGGGCCTCGATTGTAAGTAG |
| SS10 | TTGGCTCTTCTCCTTAGTAT | CATTTGTATCTTGTGAACGT |
| SS3 | CCAATGTTTGCTCCAGAT | TTCAATGACCCACGTCCC |
| SS4 | TGGTTTTCCTTGTTGCTG | GCTTGCGGCTCTGCTTAC |
| SS20 | TACAGGTATGCTGCTTTTCC | CTGGTCCTTTTCATTCTAAC |
| SS21 | AAAACATGCTCCAACAGCCT | CCAAATGTAGCCAGTGAGGA |
| SS22 | GGAGGAGTTCATTTGAGGCG | CTGGGTGGGCTAGGAAGTAA |
| SS23 | ATACCTCCTTGTATTCGCACT | CGATCGATTGCCACATTATA |
| SS24 | AGCGTGAATCTAATAGCACT | CGTTCAACAAGACCCAATAC |
| SS25 | TTGTAACCGTCGATTTCGTTC | CCGCTCCGTCACTCTACTACC |
| SS16 | CCTCCGACCTCAGCACCTGC | GTTGGCGTCCGCTGCTCCTG |
| SS17 | AGGTAGGCGTGGCGATCAAC | CTTCTCCGGTCACCATCCAC |
| SS27 | TTGGCGATTAATGATCCGGGAAC | CGTTCGTGCCGGTGATGTCG |
| SS29 | ACAACAGTTCTTCACCAGAG | GTAGTATAAATTGTAATAGCTCAA |
| SS32 | GTTAAATGAATCATCAGGAT | AGTAGTCTTGAATTCGCTGT |

Fig.1 Effects of SS2 mutation on the growth performance of seedlings under normal and salinity stress A:Growth performance of WT and ss2 mutants seedling under normal or NaCl treatment. Yellow bar = 1 cm. White bar = 5 cm. B-E:Shoot height(B),shoot fresh weight(C),Na+ concentration([Na])(D)and[Na]/[K]ratio in the shoots(E)of the ss2 mutants under normal and salinity stress conditions. The values are means ± SE of 5 replicates. Significant differences at P < 0.05 are indicated with different letters,the same below. DW:Dry weight
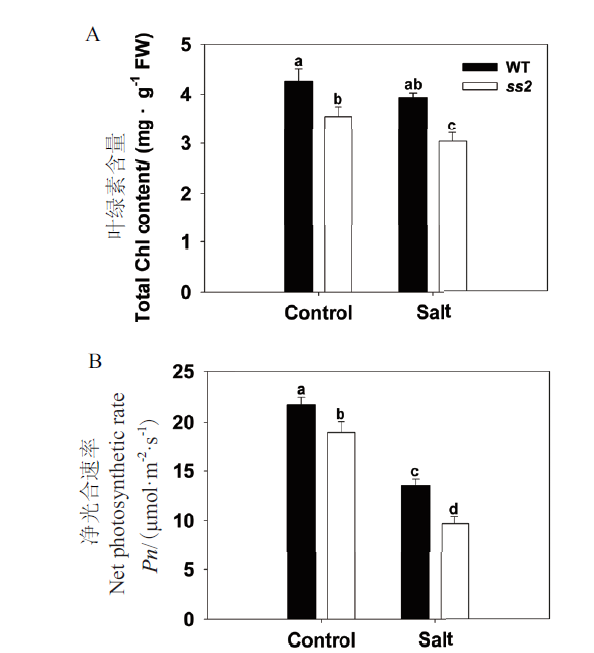
Fig. 2 Physiological responses of WT and ss2 to salinity stress during tillering stage A:Total chlorophyll content. B:Net photosynthetic rate of the leaf. FW:Fresh weight,the same below
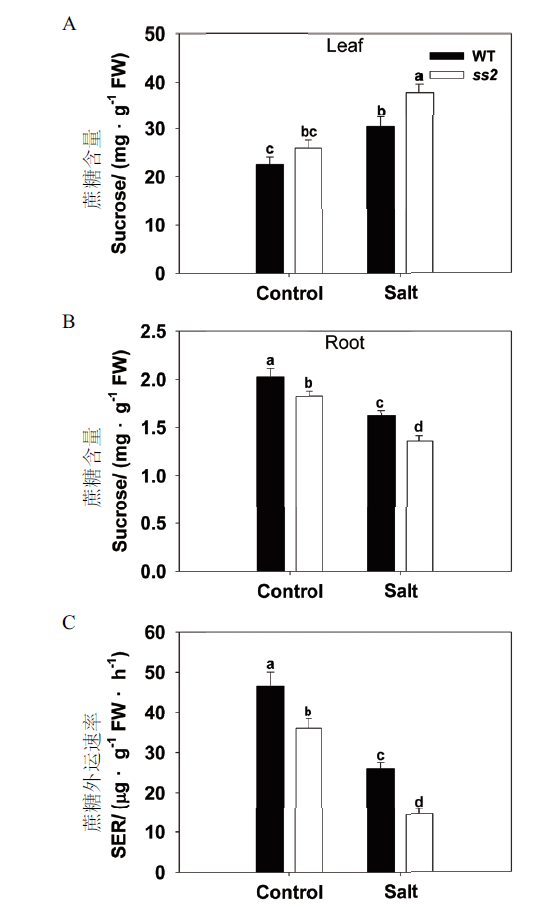
Fig. 3 Comparison of sucrose transportation between WT and ss2 in response to salinity stress A:Sucrose contents of the leaf. B:Sucrose content of the root. C:Rate of sucrose export from the leaf(SER)
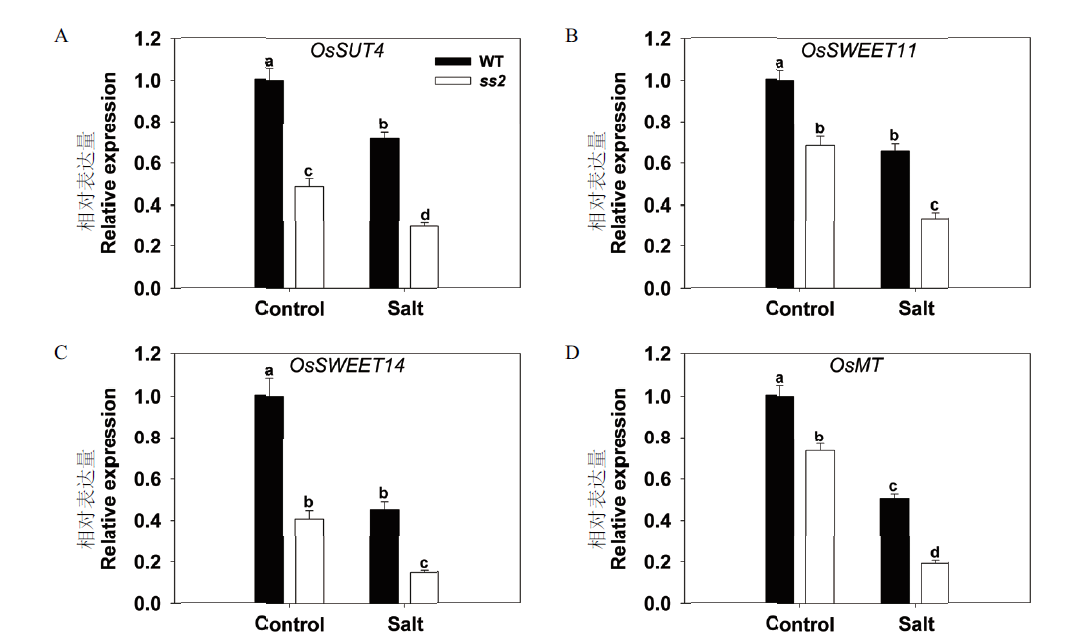
Fig.4 Effects of SS2 mutation on the expressions of genes related to sugar transport in WT and ss2 leaves The genes assayed were OsSUT4(A),OsSWEET11(B),OsSWEET14(C)and OsMT(D). UBQ5 was chosen as the reference sequence. The expression of to-be-detected genes in non-stressed WT plants was set to 1
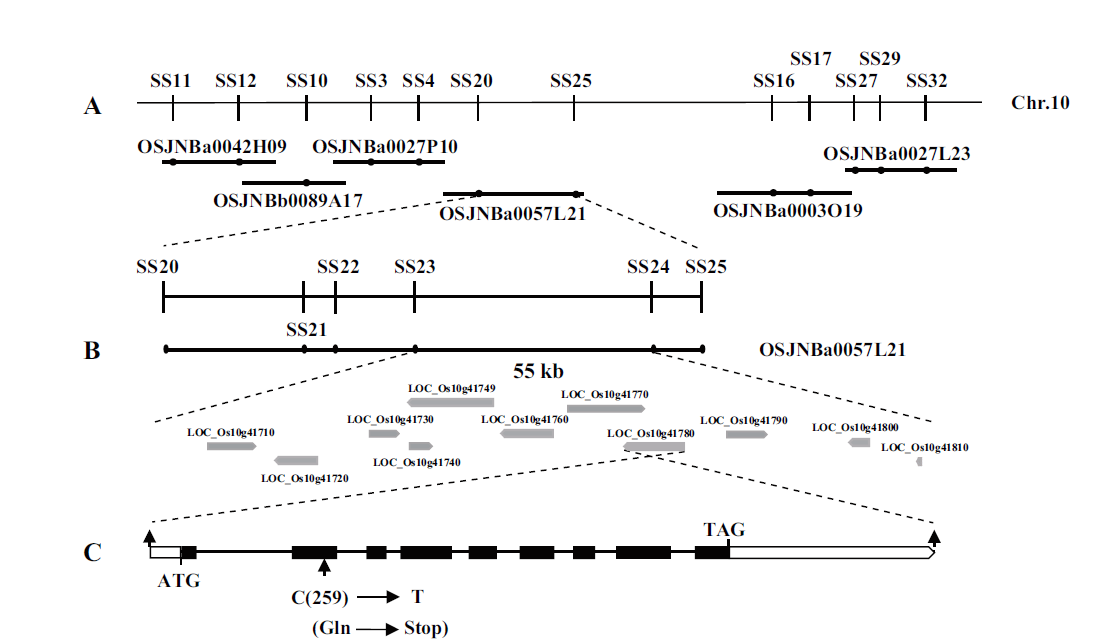
Fig.5 Map-based cloning of SS2 A:SS2 was primarily mapped on chromosome 5 and fine mapping with the markers developed based on the sequence of BAC clone OSJNBa0057L21. B:The SS2 locus was narrowed to a 55 kb genomic DNA region between markers SS23 and SS24. Eleven open reading frames were located in the region. C:Gene structure of the SS2 candidate LOC_Os10g41780. Arrows show mutation sites of ss2

Fig.6 Difference analysis in the growth performance of complementary transgenic plant compared with WT in response to salinity stress A:Growth performance of WT and complementary transgenic plants under normal or 100 mmol/L NaCl treatment. Yellow bar = 1 cm. White bar = 5 cm. B-E:Shoot biomass(B),root biomass(C),H2O2(D)and MDA(E)content in the shoots of the seedlings under normal and salinity stress conditions
| [1] |
Campbell MT, Bandillo N, Al Shiblawi FRA, et al. Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice(Oryza sativa)for root sodium content[J]. PLoS Genet, 2017, 13(6):e1006823.
doi: 10.1371/journal.pgen.1006823 URL |
| [2] |
Khan I, Khan S, Zhang Y, et al. CRISPR-Cas technology based genome editing for modification of salinity stress tolerance responses in rice(Oryza sativa L.)[J]. Mol Biol Rep, 2021, 48(4):3605-3615.
doi: 10.1007/s11033-021-06375-0 URL |
| [3] |
Machado R, Serralheiro R. Soil salinity:effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization[J]. Horticulturae, 2017, 3(2):30.
doi: 10.3390/horticulturae3020030 URL |
| [4] |
Qin H, Li YX, Huang RF. Advances and challenges in the breeding of salt-tolerant rice[J]. Int J Mol Sci, 2020, 21(21):8385.
doi: 10.3390/ijms21218385 URL |
| [5] | 刘奕媺, 于洋, 方军. 盐碱胁迫及植物耐盐碱分子机制研究[J]. 土壤与作物, 2018, 7(2):201-211. |
| Liu YM, Yu Y, Fang J. Saline-alkali stress and molecular mechanism of saline-alkali tolerance in plants[J]. Soils Crops, 2018, 7(2):201-211. | |
| [6] |
Lim JD, Cho JI, Park YI, et al. Sucrose transport from source to sink seeds in rice[J]. Physiol Plant, 2006, 126(4):572-584.
doi: 10.1111/j.1399-3054.2006.00654.x URL |
| [7] |
Eom JS, Choi SB, Ward JM, et al. The mechanism of phloem loading in rice(Oryza sativa)[J]. Mol Cells, 2012, 33(5):431-438.
doi: 10.1007/s10059-012-0071-9 URL |
| [8] |
Kerepesi I, Galiba G. Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings[J]. Crop Sci, 2000, 40(2):482-487.
doi: 10.2135/cropsci2000.402482x URL |
| [9] |
Watanabe S, Kojima K, Ide Y, et al. Effects of saline and osmotic stress on proline and sugar accumulation in Populus euphratica in vitro[J]. Plant Cell Tissue Organ Cult, 2000, 63(3):199-206.
doi: 10.1023/A:1010619503680 URL |
| [10] |
Feng G, Zhang FS, Li XL, et al. Improved tolerance of maize plants to salt stress by arbuscular mycorrhiza is related to higher accumulation of soluble sugars in roots[J]. Mycorrhiza, 2002, 12(4):185-190.
pmid: 12189473 |
| [11] |
Mathan J, Singh A, Ranjan A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice[J]. Physiol Plant, 2021, 171(4):620-637.
doi: 10.1111/ppl.13210 URL |
| [12] |
Chen G, Feng HM, Hu QD, et al. Improving rice tolerance to potassium deficiency by enhancing OsHAK16p:WOX11-controlled root development[J]. Plant Biotechnol J, 2015, 13(6):833-848.
doi: 10.1111/pbi.12320 URL |
| [13] |
Chen G, Liu CL, Gao ZY, et al. Variation in the abundance of OsHAK1 transcript underlies the differential salinity tolerance of an indica and a japonica rice cultivar[J]. Front Plant Sci, 2018, 8:2216.
doi: 10.3389/fpls.2017.02216 URL |
| [14] |
Chen G, Liu CL, Gao ZY, et al. OsHAK1, a high-affinity potassium transporter, positively regulates responses to drought stress in rice[J]. Front Plant Sci, 2017, 8:1885.
doi: 10.3389/fpls.2017.01885 pmid: 29163608 |
| [15] |
Chen G, Hu J, Dong LL, et al. The tolerance of salinity in rice requires the presence of a functional copy of FLN2[J]. Biomolecules, 2019, 10(1):17.
doi: 10.3390/biom10010017 URL |
| [16] |
Chen G, Wu C, He L, et al. Knocking out the gene RLS1 induces hypersensitivity to oxidative stress and premature leaf senescence in rice[J]. Int J Mol Sci, 2018, 19(10):2853.
doi: 10.3390/ijms19102853 URL |
| [17] |
Chen G, Hu J, Lian J, et al. Functional characterization of OsHAK1 promoter in response to osmotic/drought stress by deletion analysis in transgenic rice[J]. Plant Growth Regul, 2019, 88(3):241-251.
doi: 10.1007/s10725-019-00504-3 URL |
| [18] |
Lee S, Kim JH, Yoo ES, et al. Differential regulation of chlorophyll a oxygenase genes in rice[J]. Plant Mol Biol, 2005, 57(6):805-818.
pmid: 15952067 |
| [19] |
Yang YL, Xu J, Huang LC, et al. PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice[J]. J Exp Bot, 2016, 67(5):1297-1310.
doi: 10.1093/jxb/erv529 pmid: 26709310 |
| [20] |
Ferrandino A, Lovisolo C. Abiotic stress effects on grapevine(Vitis vinifera L.):focus on abscisic acid-mediated consequences on secondary metabolism and berry quality[J]. Environ Exp Bot, 2014, 103:138-147.
doi: 10.1016/j.envexpbot.2013.10.012 URL |
| [21] |
Aoki N, Hirose T, Scofield GN, et al. The sucrose transporter gene family in rice[J]. Plant Cell Physiol, 2003, 44(3):223-232.
pmid: 12668768 |
| [22] |
Gong X, Liu ML, Zhang LJ, et al. Arabidopsis AtSUC2 and AtSUC4, encoding sucrose transporters, are required for abiotic stress tolerance in an ABA-dependent pathway[J]. Physiol Plant, 2015, 153(1):119-136.
doi: 10.1111/ppl.12225 pmid: 24814155 |
| [23] |
Jia WQ, Zhang LJ, Wu D, et al. Sucrose transporter AtSUC9 mediated by a low sucrose level is involved in Arabidopsis abiotic stress resistance by regulating sucrose distribution and ABA accumulation[J]. Plant Cell Physiol, 2015, 56(8):1574-1587.
doi: 10.1093/pcp/pcv082 URL |
| [24] |
Ma QJ, Sun MH, Kang H, et al. A CIPK protein kinase targets sucrose transporter MdSUT2. 2 at Ser254 for phosphorylation to enhance salt tolerance[J]. Plant Cell Environ, 2019, 42(3):918-930.
doi: 10.1111/pce.13349 URL |
| [25] |
Ma QJ, Sun MH, Lu J, et al. An apple sucrose transporter MdSUT2.2is a phosphorylation target for protein kinase MdCIPK22 in response to drought[J]. Plant Biotechnol J, 2019, 17(3):625-637.
doi: 10.1111/pbi.13003 URL |
| [26] | Ibraheem O, Dealtry G, Roux S, et al. The effect of drought and salinity on the expressional levels of sucrose transporters in rice(Oryza sativa Nipponbare)cultivar plants[J]. Plant Omics, 2011, 4(2):68-74. |
| [27] |
Siahpoosh MR, Sanchez DH, Schlereth A, et al. Modification of OsSUT1 gene expression modulates the salt response of rice Oryza sativa cv. Taipei 309[J]. Plant Sci, 2012, 182:101-111.
doi: 10.1016/j.plantsci.2011.01.001 pmid: 22118621 |
| [28] |
Zhou AM, Ma HP, Feng S, et al. A novel sugar transporter from Dianthus spiculifolius, DsSWEET12, affects sugar metabolism and confers osmotic and oxidative stress tolerance in Arabidopsis[J]. Int J Mol Sci, 2018, 19(2):497.
doi: 10.3390/ijms19020497 URL |
| [29] |
Zhou AM, Ma HP, Feng S, et al. DsSWEET17, a tonoplast-localized sugar transporter from Dianthus spiculifolius, affects sugar metabolism and confers multiple stress tolerance in Arabidopsis[J]. Int J Mol Sci, 2018, 19(6):1564.
doi: 10.3390/ijms19061564 URL |
| [30] |
Platten JD, Egdane JA, Ismail AM. Salinity tolerance, Na+ exclusion and allele mining of HKT1;5 in Oryza sativa and O. glaberrima:many sources, many genes, one mechanism?[J]. BMC Plant Biol, 2013, 13:32.
doi: 10.1186/1471-2229-13-32 pmid: 23445750 |
| [31] |
Yeo AR, Yeo ME, Flowers SA, et al. Screening of rice(Oryza sativa L.)genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance[J]. Theor Appl Genet, 1990, 79(3):377-384.
doi: 10.1007/BF01186082 pmid: 24226357 |
| [32] |
Rahman MA, Thomson MJ, Shah-E-Alam M, et al. Exploring novel genetic sources of salinity tolerance in rice through molecular and physiological characterization[J]. Ann Bot, 2016, 117(6):1083-1097.
doi: 10.1093/aob/mcw030 URL |
| [1] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [2] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [3] | YAO Sha-sha, WANG Jing-jing, WANG Jun-jie, LIANG Wei-hong. Molecular Mechanisms of Rice Grain Size Regulation Related to Plant Hormone Signaling Pathways [J]. Biotechnology Bulletin, 2023, 39(8): 80-90. |
| [4] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [5] | LIANG Cheng-gang, WANG Yan, LI Tian, OHSUGI Ryu, AOKI Naohiro. Effect of SP1 on Panicle Architecture by Regulating Carbohydrate Remobilization [J]. Biotechnology Bulletin, 2023, 39(5): 152-159. |
| [6] | ZHOU Ding-ding, LI Hui-hu, TANG Xing-yong, YU Fa-xin, KONG Dan-yu, LIU Yi. Research Progress in the Biosynthesis and Regulation of Glycyrrhizic Acid and Liquiritin [J]. Biotechnology Bulletin, 2023, 39(5): 44-53. |
| [7] | LI Wang-ning, ZHANG Hao-jie, LI Ya-nan, LIANG Meng-jing, JI Chun-li, Zhang Chun-hui, LI Run-zhi, CUI Yu-lin, QIN Song, CUI Hong-li. Phenotypic Characterization of Blue Photoreceptor Plant Type Cryptochrome CRY Mutant in Chlamydomonas reinhardtii [J]. Biotechnology Bulletin, 2023, 39(2): 243-253. |
| [8] | YANG Mao, LIN Yu-feng, DAI Yang-shuo, PAN Su-jun, PENG Wei-ye, YAN Ming-xiong, LI Wei, WANG Bing, DAI Liang-ying. OsDIS1 Negatively Regulates Rice Drought Tolerance Through Antioxidant Pathways [J]. Biotechnology Bulletin, 2023, 39(2): 88-95. |
| [9] | JIANG Min-xuan, LI Kang, LUO Liang, LIU Jian-xiang, LU Hai-ping. Advances on the Expressions of Foreign Proteins in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 110-122. |
| [10] | JIANG Nan, SHI Yang, ZHAO Zhi-hui, LI Bin, ZHAO Yi-hui, YANG Jun-biao, YAN Jia-ming, JIN Yu-fan, CHEN Ji, HUANG Jin. Expression and Functional Analysis of OsPT1 Gene in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2023, 39(1): 166-174. |
| [11] | GAO Xiao-rong, DING Yao, LV Jun. Effects of Pseudomonas sp. PR3,a Pyrene-degrading Bacterium with Plant Growth-promoting Properties,on Rice Growth Under Pyrene Stress [J]. Biotechnology Bulletin, 2022, 38(9): 226-236. |
| [12] | HUANG Jing, ZHU Liang, XUE Peng-bo, FU Qiang. Research on Mechanism and QTL Mapping Associated with Cadmium Accumulation in Rice Leaves and Grains [J]. Biotechnology Bulletin, 2022, 38(8): 118-126. |
| [13] | TANG Guang-fu, GUI Yan-ling, MAN Hai-qiao, ZHAO Jie-hong. Editing pyrG Gene of Monascus by CRISPR/Cas 9 and Its Effects on Secondary Metabolism [J]. Biotechnology Bulletin, 2022, 38(8): 198-205. |
| [14] | CHEN Guang, LI Jia, DU Rui-ying, WANG Xu. pOsHAK1:OsFLN2 Expression Enhances the Drought Tolerance by Altering Sugar Metabolism in Rice [J]. Biotechnology Bulletin, 2022, 38(8): 92-100. |
| [15] | LI Bai, CAI Zhi-jun, WANG Lei, CHEN Jie, CAO Kui-rong, LI Jun, CHONG Gao-jun. Development and Application of the Combinatorial Marker for the Rice Blast Resistance Gene Pigm [J]. Biotechnology Bulletin, 2022, 38(7): 153-159. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||