








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (11): 129-139.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0077
Previous Articles Next Articles
LAI Gong-ti1,2( ), QUE Qiu-xia1, PAN Ruo1, LIU Yu-xuan1, WANG Qi1,2, LAI Pu-fu1, GAO Hui-ying1,2, LAI Cheng-chun1,2(
), QUE Qiu-xia1, PAN Ruo1, LIU Yu-xuan1, WANG Qi1,2, LAI Pu-fu1, GAO Hui-ying1,2, LAI Cheng-chun1,2( )
)
Received:2021-01-18
Online:2022-11-26
Published:2022-12-01
Contact:
LAI Cheng-chun
E-mail:laigt4769@163.com;lccisland@163.com
LAI Gong-ti, QUE Qiu-xia, PAN Ruo, LIU Yu-xuan, WANG Qi, LAI Pu-fu, GAO Hui-ying, LAI Cheng-chun. Response of Chalcone Synthase Gene(CHS)to Different Light Quality and Transcription Factor Regulation in Vitis davidii[J]. Biotechnology Bulletin, 2022, 38(11): 129-139.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Application |
|---|---|---|
| CHS2-F | CACTGATTCAAGCAGCCAAAAATG | CHS2 克隆 |
| CHS2-R | TTACTTAAGTGTGAAAGGAGAGAAG | |
| CHS3-F | CTCCAACCACCACAACCCTCATC | CHS3 克隆 |
| CHS3-R | ATGGGGCTAGAAAAAGCATATCATC | |
| CHS2-qPCR-F | TGGGTCTGAAGGAAGAGAAAC | CHS2 qPCR |
| CHS2-qPCR-R | TTGTGTAGCAAGGCTGTGC | |
| CHS3-qPCR-F | AGTTACGCTCCACACGACAC | CHS3 qPCR |
| CHS3-qPCR-R | AGCACCACGGTCTCAACAGT |
Table 1 Primer information
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Application |
|---|---|---|
| CHS2-F | CACTGATTCAAGCAGCCAAAAATG | CHS2 克隆 |
| CHS2-R | TTACTTAAGTGTGAAAGGAGAGAAG | |
| CHS3-F | CTCCAACCACCACAACCCTCATC | CHS3 克隆 |
| CHS3-R | ATGGGGCTAGAAAAAGCATATCATC | |
| CHS2-qPCR-F | TGGGTCTGAAGGAAGAGAAAC | CHS2 qPCR |
| CHS2-qPCR-R | TTGTGTAGCAAGGCTGTGC | |
| CHS3-qPCR-F | AGTTACGCTCCACACGACAC | CHS3 qPCR |
| CHS3-qPCR-R | AGCACCACGGTCTCAACAGT |
| 基因名称 Gene name | GenBank 登陆号 GenBank accession | 片段长度 Fragment length/bp | ORF长度 ORF length/bp | 氨基酸数目 Amino acid number | 分子量 Molecular weight/kD | 理论等电点pI | 亲水性 GRAVY | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| VdCHS2 | OL906400 | 1 382 | 1 182 | 393 | 42.90 | 6.1 | -0.057 | 无 No | 细胞质Cytoplasmic |
| VdCHS3 | OL906401 | 1 398 | 1 170 | 389 | 42.60 | 6.18 | -0.132 | 无 No | 细胞质Cytoplasmic |
Table 2 Bioinformatics analysis of VdCHS
| 基因名称 Gene name | GenBank 登陆号 GenBank accession | 片段长度 Fragment length/bp | ORF长度 ORF length/bp | 氨基酸数目 Amino acid number | 分子量 Molecular weight/kD | 理论等电点pI | 亲水性 GRAVY | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| VdCHS2 | OL906400 | 1 382 | 1 182 | 393 | 42.90 | 6.1 | -0.057 | 无 No | 细胞质Cytoplasmic |
| VdCHS3 | OL906401 | 1 398 | 1 170 | 389 | 42.60 | 6.18 | -0.132 | 无 No | 细胞质Cytoplasmic |
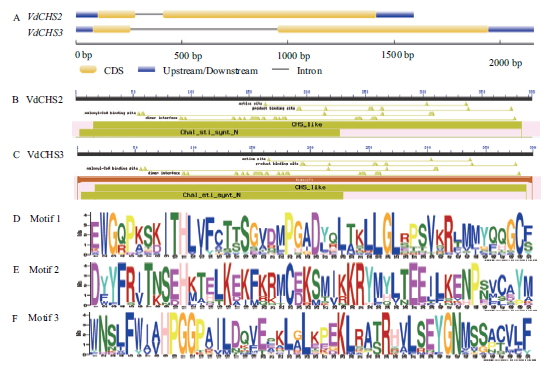
Fig.1 Gene structure and conserved domain of VdCHS2 and VdCHS3 A:Exon/intron structure of VdCHS2 and VdCHS3. B and C:Conserved domain of VdCHS2 and VdCHS3. D-F:Motifs of CHS proteins
| 光质类型 Light quality type | 光质名称 Light quality | 光波长 Light wavelength/nm | 表达模式 Expression pattern | ||
|---|---|---|---|---|---|
| 主峰 Main peak | 次峰 Secondary peak | VdCHS2 | VdCHS3 | ||
| 短波光 | 紫光 | 448 | / | 先升后降 | 先升后降 |
| 蓝光 | 456 | / | 先升后降 | 先升后降 | |
| 绿光 | 514 | / | 先升后降 | 先升后降 | |
| 长波光 | 黄光 | 590 | / | 下调 | 下调 |
| 红光 | 637 | / | 下调 | 下调 | |
| 混合光质 | 白光 | 453 | 550 | 先升后降 | 下调 |
| 暖白光 | 588 | 450 | 先升后降 | 上调 | |
| 暖黄光 | 595 | 450 | 先升后降 | 先升后降 | |
Table 3 Types of light quality and VdCHS expression model
| 光质类型 Light quality type | 光质名称 Light quality | 光波长 Light wavelength/nm | 表达模式 Expression pattern | ||
|---|---|---|---|---|---|
| 主峰 Main peak | 次峰 Secondary peak | VdCHS2 | VdCHS3 | ||
| 短波光 | 紫光 | 448 | / | 先升后降 | 先升后降 |
| 蓝光 | 456 | / | 先升后降 | 先升后降 | |
| 绿光 | 514 | / | 先升后降 | 先升后降 | |
| 长波光 | 黄光 | 590 | / | 下调 | 下调 |
| 红光 | 637 | / | 下调 | 下调 | |
| 混合光质 | 白光 | 453 | 550 | 先升后降 | 下调 |
| 暖白光 | 588 | 450 | 先升后降 | 上调 | |
| 暖黄光 | 595 | 450 | 先升后降 | 先升后降 | |
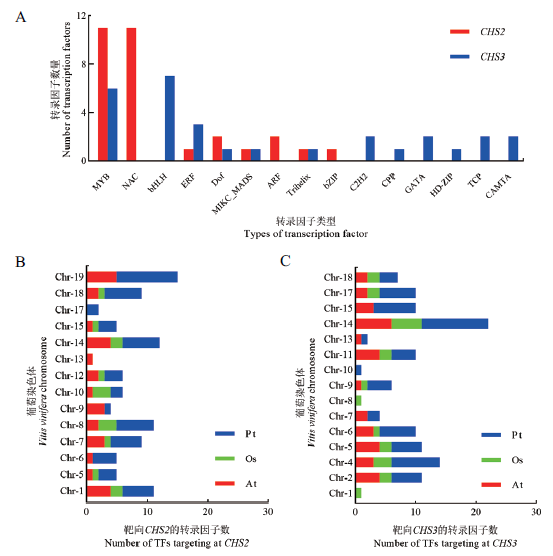
Fig.5 Transcription factors targeting at CHS and collinea-rity analysis in multiple species A:Type and number of transcription factors targeting at CHS. B and C:Number of collinearity transcription factors targeting at CHS2/CHS3 on each chromosome of grapevine with other species. At:Arabidopsis thaliana. Os:Oryza sativa. Pt:Populus trichocarpa. The same below
| MYB基因ID MYB gene ID | 注释 Annotation | 染色体 Chromosome | 起始位点 Start | 终止位点 End | 正负链 Strand | 基因长度 Gene length/bp | ORF长度 ORF length/bp | 氨基酸数 Amino acid number | 靶基因 Target gene |
|---|---|---|---|---|---|---|---|---|---|
| VIT_00s1352g00010 | MYB-RAX3 | Unrandom | 38 747 500 | 38 748 788 | - | 1 057 | 1 020 | 339 | CHS2/CHS3 |
| VIT_05s0049g01020 | MYB4 | Chr-5 | 8 046 871 | 8 048 411 | + | 1 212 | 762 | 253 | CHS2 |
| VIT_05s0077g01360 | MYB12 | Chr-5 | 1 104 556 | 1 106 597 | + | 1 152 | 1 152 | 383 | CHS2 |
| VIT_06s0004g02110 | MYB46 | Chr-6 | 2 573 382 | 2 575 343 | + | 1 017 | 978 | 325 | CHS2/CHS3 |
| VIT_07s0005g01210 | Unnamed | Chr-7 | 3 730 465 | 3 732 879 | - | 1 247 | 1 104 | 367 | CHS2 |
| VIT_07s0141g00100 | MYB4-like | Chr-7random | 56 798 | 57 999 | - | 999 | 993 | 330 | CHS2/CHS3 |
| VIT_08s0056g00800 | MYB60 | Chr-8 | 1 264 742 | 1 266 497 | + | 1 269 | 966 | 321 | CHS3 |
| VIT_14s0060g00240 | MYB86 | Chr-14 | 240 314 | 242 056 | - | 1 532 | 1 320 | 439 | CHS2/CHS3 |
| VIT_14s0108g00830 | MYB306 | Chr-14 | 29 511 863 | 29 514 026 | - | 1 312 | 984 | 327 | CHS3 |
| VIT_14s0219g00050 | MYB315-like | Chr-14 | 26 355 901 | 26 357 181 | + | 935 | 807 | 268 | CHS2 |
| VIT_15s0046g03190 | MYB308 | Chr-15 | 19 778 733 | 19 781 642 | - | 1 105 | 909 | 302 | CHS2 |
| VIT_19s0085g00050 | Unnamed | Chr-19 | 22 358 835 | 22 361 459 | + | 1 095 | 1 095 | 364 | CHS2 |
| VIT_19s0085g00940 | MYB26 | Chr-19 | 23 462 631 | 23 464 270 | + | 1 453 | 1 014 | 337 | CHS2 |
Table 4 Information of MYB transcription factors targeting at CHS
| MYB基因ID MYB gene ID | 注释 Annotation | 染色体 Chromosome | 起始位点 Start | 终止位点 End | 正负链 Strand | 基因长度 Gene length/bp | ORF长度 ORF length/bp | 氨基酸数 Amino acid number | 靶基因 Target gene |
|---|---|---|---|---|---|---|---|---|---|
| VIT_00s1352g00010 | MYB-RAX3 | Unrandom | 38 747 500 | 38 748 788 | - | 1 057 | 1 020 | 339 | CHS2/CHS3 |
| VIT_05s0049g01020 | MYB4 | Chr-5 | 8 046 871 | 8 048 411 | + | 1 212 | 762 | 253 | CHS2 |
| VIT_05s0077g01360 | MYB12 | Chr-5 | 1 104 556 | 1 106 597 | + | 1 152 | 1 152 | 383 | CHS2 |
| VIT_06s0004g02110 | MYB46 | Chr-6 | 2 573 382 | 2 575 343 | + | 1 017 | 978 | 325 | CHS2/CHS3 |
| VIT_07s0005g01210 | Unnamed | Chr-7 | 3 730 465 | 3 732 879 | - | 1 247 | 1 104 | 367 | CHS2 |
| VIT_07s0141g00100 | MYB4-like | Chr-7random | 56 798 | 57 999 | - | 999 | 993 | 330 | CHS2/CHS3 |
| VIT_08s0056g00800 | MYB60 | Chr-8 | 1 264 742 | 1 266 497 | + | 1 269 | 966 | 321 | CHS3 |
| VIT_14s0060g00240 | MYB86 | Chr-14 | 240 314 | 242 056 | - | 1 532 | 1 320 | 439 | CHS2/CHS3 |
| VIT_14s0108g00830 | MYB306 | Chr-14 | 29 511 863 | 29 514 026 | - | 1 312 | 984 | 327 | CHS3 |
| VIT_14s0219g00050 | MYB315-like | Chr-14 | 26 355 901 | 26 357 181 | + | 935 | 807 | 268 | CHS2 |
| VIT_15s0046g03190 | MYB308 | Chr-15 | 19 778 733 | 19 781 642 | - | 1 105 | 909 | 302 | CHS2 |
| VIT_19s0085g00050 | Unnamed | Chr-19 | 22 358 835 | 22 361 459 | + | 1 095 | 1 095 | 364 | CHS2 |
| VIT_19s0085g00940 | MYB26 | Chr-19 | 23 462 631 | 23 464 270 | + | 1 453 | 1 014 | 337 | CHS2 |

Fig.6 Collinearity analysis of MYB targeting at the CHS in multiple species A and B:Collinearity of MYB targeting at CHS2/CHS3 between grapevine,Arabidopsis,rice and P. trichocarpa. C and D:Number of collinearity MYB targeting at CHS2/CHS3 on each chromosome of grapevine with other species
| [1] | Hatier JHB, Clearwater MJ, Gould KS. The functional significance of black-pigmented leaves:photosynthesis, photoprotection and productivity in Ophiopogon planiscapus ‘Nigrescens’[J]. PLoS One, 2013, 8(6):e67850. |
| [2] |
Ju YL, Yue XF, Cao XY, et al. Targeted metabolomic and transcript level analysis reveals quality characteristic of Chinese wild grapes(Vitis davidii Foex)[J]. Foods, 2020, 9(10):1387.
doi: 10.3390/foods9101387 URL |
| [3] |
Fu YF, Chen M, Ye XL, et al. Variation laws of anthocyanin content in roots and their relationships with major economic traits in purple-fleshed sweetpotato[Ipomoea batatas(L.)Lam][J]. Agric Sci China, 2008, 7(1):32-40.
doi: 10.1016/S1671-2927(08)60019-X URL |
| [4] | 刘恺媛, 王茂良, 辛海波, 等. 植物花青素合成与调控研究进展[J]. 中国农学通报, 2021, 37(14):41-51. |
| Liu KY, Wang ML, Xin HB, et al. Anthocyanin biosynthesis and regulate mechanisms in plants:a review[J]. Chin Agric Sci Bull, 2021, 37(14):41-51. | |
| [5] |
Wang XC, Wu J, Guan ML, et al. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis[J]. Plant J, 2020, 101(3):637-652.
doi: 10.1111/tpj.14570 URL |
| [6] |
Yonekura-Sakakibara K, Higashi Y, Nakabayashi R. The origin and evolution of plant flavonoid metabolism[J]. Front Plant Sci, 2019, 10:943.
doi: 10.3389/fpls.2019.00943 pmid: 31428108 |
| [7] | 周小理, 王青, 等. 植物中查尔酮酶合成黄酮类化合物的研究进展[J]. 食品工业, 2009, 30(6):72-74. |
| Zhou XL, Wang Q, et al. Advances of plant chalcone synthase in synthesis of flavonoids[J]. Food Ind, 2009, 30(6):72-74. | |
| [8] | 苏全胜, 王爽, 孙玉强, 等. 植物原花青素生物合成及调控研究进展[J]. 中国细胞生物学学报, 2021, 43(1):219-229. |
| Su QS, Wang S, Sun YQ, et al. Advances in biosynthesis and regulation of plant proanthocyanidins[J]. Chin J Cell Biol, 2021, 43(1):219-229. | |
| [9] | 胡可, 韩科厅, 戴思兰. 环境因子调控植物花青素苷合成及呈色的机理[J]. 植物学报, 2010, 45(3):307-317. |
| Hu K, Han KT, Dai SL. Regulation of plant anthocyanin synthesis and pigmentation by environmental factors[J]. Chin Bull Bot, 2010, 45(3):307-317. | |
| [10] | 潘红, 赖呈纯, 黄贤贵, 等. 不同处理对刺葡萄愈伤组织花青素和原花青素生物合成的影响[J]. 热带作物学报, 2018, 39(12):2404-2409. |
| Pan H, Lai CC, Huang XG, et al. Effects of different treatments on anthocyanins and procyanidins biosynthesis in spine grape callus[J]. Chin J Trop Crops, 2018, 39(12):2404-2409. | |
| [11] |
卢雯瑩, 赵磊, 李天奇, 等. 蔷薇科植物果实花青苷积累研究进展[J]. 生物技术通报, 2021, 37(1):234-245.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0717 |
| Lu WY, Zhao L, Li TQ, et al. Research advances of fruit anthocyanin accumulation in Rosaceae plants[J]. Biotechnol Bull, 2021, 37(1):234-245. | |
| [12] | 崔虎亮, 贺霞, 张前. 不同牡丹品种开花期间花瓣花青素和类黄酮组成的动态变化[J]. 中国农业科学, 2021, 54(13):2858-2869. |
| Cui HL, He X, Zhang Q. Anthocyanins and flavonoids accumulation forms of five different color tree peony cultivars at blooming stages[J]. Sci Agric Sin, 2021, 54(13):2858-2869. | |
| [13] |
Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity[J]. Trends Plant Sci, 2005, 10(2):63-70.
pmid: 15708343 |
| [14] |
Mishra AK, Puranik S, Prasad M. Structure and regulatory networks of WD40 protein in plants[J]. J Plant Biochem Biotechnol, 2012, 21(1):32-39.
doi: 10.1007/s13562-012-0134-1 URL |
| [15] |
Kui LW, Bolitho K, Grafton K, et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae[J]. BMC Plant Biol, 2010, 10:50.
doi: 10.1186/1471-2229-10-50 URL |
| [16] |
Bogs J, Jaffé FW, et al. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development[J]. Plant Physiol, 2007, 143(3):1347-1361.
pmid: 17208963 |
| [17] |
Walker AR, Lee E, Bogs J, et al. White grapes arose through the mutation of two similar and adjacent regulatory genes[J]. Plant J, 2007, 49(5):772-785.
pmid: 17316172 |
| [18] |
Spelt C, Quattrocchio F, Mol JN, et al. anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes[J]. Plant Cell, 2000, 12(9):1619-1632.
pmid: 11006336 |
| [19] |
Jin HL, Cominelli E, Bailey P, et al. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis[J]. EMBO J, 2000, 19(22):6150-6161.
pmid: 11080161 |
| [20] | 张静, 潘红, 赖呈纯, 等. ALA对刺葡萄愈伤组织生长及主要抗氧化物质积累的影响[J]. 热带作物学报, 2021, 42(8):2305-2312. |
| Zhang J, Pan H, Lai CC, et al. Effect of 5-aminolevulinic acid (ALA)on callus growth of spine grape callus and accumulation of its main antioxidants[J]. Chin J Trop Crops, 2021, 42(8):2305-2312. | |
| [21] |
Ju YL, Yang L, Yue XF, et al. Anthocyanin profiles and color properties of red wines made from Vitis davidii and Vitis vinifera grapes[J]. Food Sci Hum Wellness, 2021, 10(3):335-344.
doi: 10.1016/j.fshw.2021.02.025 URL |
| [22] |
王静, 周广胜. 中国毛葡萄和刺葡萄分布的气候适宜性[J]. 应用生态学报, 2020, 31(1):97-103.
doi: 10.13287/j.1001-9332.202001.019 |
| Wang J, Zhou GS. Climatic suitability for the distribution of Vitis heyneana and V. davidii in China[J]. Chin J Appl Ecol, 2020, 31(1):97-103. | |
| [23] | 洪艳, 武宇薇, 宋想, 等. 光照调控园艺作物花青素苷生物合成的分子机制[J]. 园艺学报, 2021, 48(10):1983-2000. |
| Hong Y, Wu YW, Song X, et al. Molecular mechanism of light-induced anthocyanin biosynthesis in horticultural crops[J]. Acta Hortic Sin, 2021, 48(10):1983-2000. | |
| [24] | 刘帅, 张亚红, 徐伟荣, 等. 基于转录组研究光质对转色期红地球葡萄果实着色及品质的影响[J]. 果树学报, 2021, 38(12):2045-2058. |
| Liu S, Zhang YH, Xu WR, et al. Effects of light quality on the berry coloration and quality of Red Globe grape during veraison based on transcriptome sequencing[J]. J Fruit Sci, 2021, 38(12):2045-2058. | |
| [25] | 袁华招, 赵密珍, 吴伟民, 等. 葡萄CHS和STS基因家族生物信息学鉴定和表达分析[J]. 植物遗传资源学报, 2016, 17(4):756-765. |
| Yuan HZ, Zhao MZ, Wu WM, et al. Genome-wide identification and expression analysis of CHS and STS gene families in grape(Vitis vinifera L.)[J]. J Plant Genet Resour, 2016, 17(4):756-765. | |
| [26] | 潘红, 赖呈纯, 张静, 等. 不同光质条件下刺葡萄红色愈伤组织的RT-qPCR内参基因筛选[J]. 应用与环境生物学报, 2019, 25(6):1407-1413. |
| Pan H, Lai CC, Zhang J, et al. Selection of reference genes for RT-qPCR from the red callus of Vitis davidii(Rom. CailL.)Foëx under different light qualities[J]. Chin J Appl Environ Biol, 2019, 25(6):1407-1413. | |
| [27] | 葛翠莲, 黄春辉, 徐小彪. 果实花青素生物合成研究进展[J]. 园艺学报, 2012, 39(9):1655-1664. |
| Ge CL, Huang CH, Xu XB. Research on anthocyanins biosynthesis in fruit[J]. Acta Hortic Sin, 2012, 39(9):1655-1664. | |
| [28] | 时晓芳, 林玲, 白先进, 等. 阳光玫瑰葡萄果实生长发育及品质对不同光质的响应[J]. 南方农业学报, 2021, 52(6):1641-1647. |
| Shi XF, Lin L, Bai XJ, et al. Response of berries development and quality of Shine Muscat grape to different light qualities[J]. J South Agric, 2021, 52(6):1641-1647. | |
| [29] |
张克坤, 刘凤之, 王孝娣, 等. 不同光质补光对促早栽培‘瑞都香玉’葡萄果实品质的影响[J]. 应用生态学报, 2017, 28(1):115-126.
doi: 10.13287/j.1001-9332.201701.003 |
| Zhang KK, Liu FZ, Wang XD, et al. Effects of supplementary light with different wavelengths on fruit quality of ‘Ruidu Xiangyu’ grape under promoted cultivation[J]. Chin J Appl Ecol, 2017, 28(1):115-126. | |
| [30] |
Li Q, Kubota C. Effects of supplemental light quality on growth and phytochemicals of baby leaf lettuce[J]. Environ Exp Bot, 2009, 67(1):59-64.
doi: 10.1016/j.envexpbot.2009.06.011 URL |
| [31] |
Ren J, Guo SS, Xu CL, et al. Effects of different carbon dioxide and LED lighting levels on the anti-oxidative capabilities of Gynura bicolor DC[J]. Adv Space Res, 2014, 53(2):353-361.
doi: 10.1016/j.asr.2013.11.019 URL |
| [32] |
Kadomura-Ishikawa Y, Miyawaki K, Noji S, et al. Phototropin 2 is involved in blue light-induced anthocyanin accumulation in Fragaria x ananassa fruits[J]. J Plant Res, 2013, 126(6):847-857.
doi: 10.1007/s10265-013-0582-2 pmid: 23982948 |
| [33] | 解潇冬, 刘晓莹, 汪文杰, 等. 光质对红阳猕猴桃愈伤组织生长速度和花青素合成的影响[J]. 山西农业科学, 2021, 49(10):1166-1172. |
| Xie XD, Liu XY, Wang WJ, et al. Effects of light quality on callus growth rate and anthocyanin synthesis of Hongyang kiwifruit[J]. J Shanxi Agric Sci, 2021, 49(10):1166-1172. | |
| [34] |
Liu CC, Chi C, Jin LJ, et al. The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato[J]. Plant Cell Environ, 2018, 41(8):1762-1775.
doi: 10.1111/pce.13171 URL |
| [35] | 王峰, 王秀杰, 赵胜男, 等. 光对园艺植物花青素生物合成的调控作用[J]. 中国农业科学, 2020, 53(23):4904-4917. |
| Wang F, Wang XJ, Zhao SN, et al. Light regulation of anthocyanin biosynthesis in horticultural crops[J]. Sci Agric Sin, 2020, 53(23):4904-4917. | |
| [36] |
Borevitz JO, Xia Y, Blount J, et al. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis[J]. Plant Cell, 2000, 12(12):2383-2394.
pmid: 11148285 |
| [37] |
Tohge T, Nishiyama Y, Hirai MY, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor[J]. Plant J, 2005, 42(2):218-235.
doi: 10.1111/j.1365-313X.2005.02371.x URL |
| [38] |
Zhang YZ, Xu SZ, Ma HP, et al. The R2R3-MYB gene PsMYB58 positively regulates anthocyanin biosynthesis in tree peony flowers[J]. Plant Physiol Biochem, 2021, 164:279-288.
doi: 10.1016/j.plaphy.2021.04.034 URL |
| [39] |
Lijavetzky D, Ruiz-García L, Cabezas JA, et al. Molecular genetics of berry colour variation in table grape[J]. Mol Genet Genomics, 2006, 276(5):427-435.
pmid: 16924546 |
| [40] |
Azuma A, Kobayashi S, Goto-Yamamoto N, et al. Color recovery in berries of grape(Vitis vinifera L.)‘Benitaka’, a bud sport of ‘Italia’, is caused by a novel allele at the VvmybA1 locus[J]. Plant Sci, 2009, 176(4):470-478.
doi: 10.1016/j.plantsci.2008.12.015 URL |
| [41] |
Azuma A, et al. Haplotype composition at the color locus is a major genetic determinant of skin color variation in Vitis × labruscana grapes[J]. Theor Appl Genet, 2011, 122(7):1427-1438.
doi: 10.1007/s00122-011-1542-7 URL |
| [42] |
Albert NW, Davies KM, et al. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots[J]. Plant Cell, 2014, 26(3):962-980.
doi: 10.1105/tpc.113.122069 URL |
| [43] | Zhang H, Gong JX, Chen KL, et al. A novel R3 MYB transcriptional repressor, MaMYBx, finely regulates anthocyanin biosynthesis in grape hyacinth[J]. Plant Sci, 2020, 298:110588. |
| [44] |
Fu ZZ, Wang LM, Shang HQ, et al. An R3-MYB gene of Phalaenopsis, MYBx1, represses anthocyanin accumulation[J]. Plant Growth Regul, 2019, 88(2):129-138.
doi: 10.1007/s10725-019-00493-3 URL |
| [45] |
Wan SZ, Li CF, Ma XD, et al. PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar[J]. Plant Cell Rep, 2017, 36(8):1263-1276.
doi: 10.1007/s00299-017-2151-y pmid: 28523445 |
| [46] |
Xu HF, Yang GX, Zhang J, et al. Overexpression of a repressor MdMYB15L negatively regulates anthocyanin and cold tolerance in red-fleshed callus[J]. Biochem Biophys Res Commun, 2018, 500(2):405-410.
doi: 10.1016/j.bbrc.2018.04.088 URL |
| [47] | Deng GM, Zhang S, et al. MaMYB4, an R2R3-MYB repressor transcription factor, negatively regulates the biosynthesis of anthocyanin in banana[J]. Front Plant Sci, 2021, 11:600704. |
| [48] |
Pérez-Díaz JR, Pérez-Díaz J, Madrid-Espinoza J, et al. New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco[J]. Plant Mol Biol, 2016, 90(1/2):63-76.
doi: 10.1007/s11103-015-0394-y URL |
| [49] |
Zhu ZG, Li GR, Liu L, et al. A R2R3-MYB transcription factor, VvMYBC2L2, functions as a transcriptional repressor of anthocyanin biosynthesis in grapevine(Vitis vinifera L.)[J]. Molecules, 2018, 24(1):92.
doi: 10.3390/molecules24010092 URL |
| [50] |
Tirumalai V, Swetha C, Nair A, et al. miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes[J]. J Exp Bot, 2019, 70(18):4775-4792.
doi: 10.1093/jxb/erz264 pmid: 31145783 |
| [51] |
Chagné D, Kui LW, Espley RV, et al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes[J]. Plant Physiol, 2013, 161(1):225-239.
doi: 10.1104/pp.112.206771 pmid: 23096157 |
| [52] |
Cui DL, Zhao SX, Xu HN, et al. The interaction of MYB, bHLH and WD40 transcription factors in red pear(Pyrus pyrifolia)peel[J]. Plant Mol Biol, 2021, 106(4/5):407-417.
doi: 10.1007/s11103-021-01160-w URL |
| [1] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [2] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [3] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [4] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [5] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [6] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [7] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [8] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [9] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [10] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [11] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| [12] | HU Ming-yue, YANG Yu, GUO Yang-dong, ZHANG Xi-chun. Functional Analysis of SlMYB96 Gene in Tomato Under Cold Stress [J]. Biotechnology Bulletin, 2023, 39(4): 236-245. |
| [13] | GE Yan-rui, ZHAO Ran, XU Jing, LI Ruo-fan, HU Yun-tao, LI Rui-li. Advances in the Development and Regulation of Vascular Cambium [J]. Biotechnology Bulletin, 2023, 39(3): 13-25. |
| [14] | LIU Cheng-xia, SUN Zong-yan, LUO Yun-bo, ZHU Hong-liang, QU Gui-qin. Multifaceted Roles of bHLH Phosphorylation in Regulation of Plant Physiological Functions [J]. Biotechnology Bulletin, 2023, 39(3): 26-34. |
| [15] | ZHAO Meng-liang, GUO Yi-ting, REN Yan-jing. Identification and Analysis of WRKY Transcription Factor Family Genes in Helianthus tuberosus [J]. Biotechnology Bulletin, 2023, 39(2): 116-125. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||