








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (3): 81-88.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0577
Previous Articles Next Articles
CAI Meng-xian( ), GAO Zuo-min, HU Li-juan, FENG Qun, WANG Hong-cheng, ZHU Bin(
), GAO Zuo-min, HU Li-juan, FENG Qun, WANG Hong-cheng, ZHU Bin( )
)
Received:2022-05-10
Online:2023-03-26
Published:2023-04-10
CAI Meng-xian, GAO Zuo-min, HU Li-juan, FENG Qun, WANG Hong-cheng, ZHU Bin. Development and Genetic Analysis of Two Nullisomic Lines(NC1 and NC2)in Natural Brassica napus[J]. Biotechnology Bulletin, 2023, 39(3): 81-88.
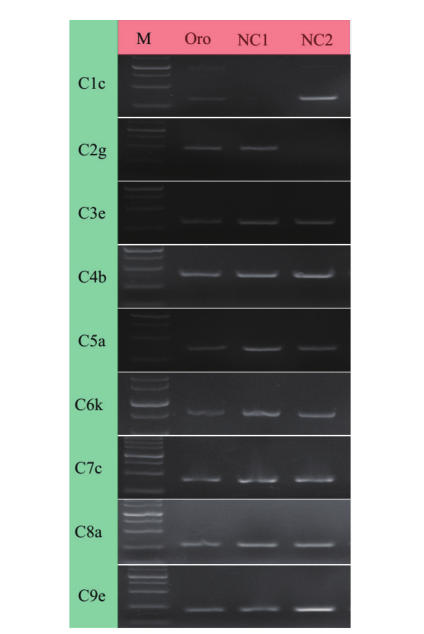
Fig. 2 Identification of C chromosome nullisomic-specific markers in NC1 and NC2 M: Marker. The mark sizes were 2 000, 1 500, 1 000, 750, 500, 250 and 100 bp. Oro is B. napus. NC1 and NC2 refer to the nullisomic of chromosome C in B. napus. C1c, C2g, …C9e are chromosomal specific primers for C genome
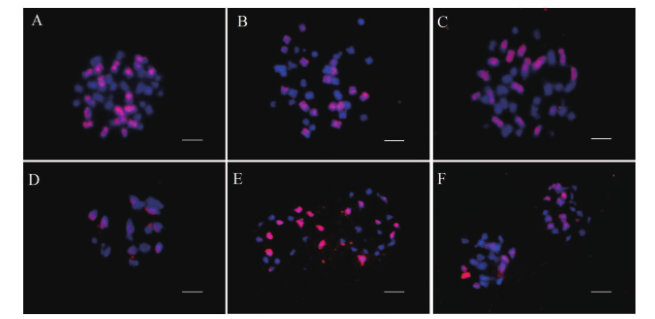
Fig. 3 FISH analysis of somatic cells and pollen mother cells of B. napus “Oro”,NC1,and NC2 A: “Oro” mitotic cells with 38 chromosomes(blue), including 18 C subgenomic chromosomes(red). B-C: NC1, NC2 mitotic cells with 36 chromosomes(blue), including 16 C subgenomic chromosomes(red). D: C2 showing 8 red hybridization signals at diakinesis. E-F: The PMC of NC2 and NC1 show equal separation of 16 hybridization signals at anaphase I. Blue indicates DAPI staining and red indicates BAC BoB014O06 probe signal. Bar: 10 μm

Fig. 4 Phenotype and pollen fertility of parental B. napus “Oro”,NC1,and NC2 A: Parental B. napus “Oro”. B: NC1 plants. The enlargement shows that blade, vein, and leaf margin cover with burrs. C: NC2 plants. The enlargement shows that NC2 plant has early squaring stage. E-F: The pollen fertility of “Oro”, NC1, and NC2

Fig. 5 Observation of chromosome behavior of at meiosis of NC1 and NC2 in B. napus A-D: Somatic chromosome number(2n=36)of NC1, PMCs of NC1 at DK, AP I, and AP II, respectively. E-H: Somatic chromosome number(2n=36)of NC2, PMCs of NC2 at DK, AP I, and AP II, respectively. I: the PMC of nullisomic show normal pairing at metaphase I. J, K: The PMCs of NC1 and NC2 with delaying chromosome at AP I. L: The PMC of NC2 with delaying chromosome at AP II. The arrows show the delaying chromosomes. Bar: 10 μm
| [1] |
Siegel JJ, Amon A. New insights into the troubles of aneuploidy[J]. Annu Rev Cell Dev Biol, 2012, 28:189-214.
doi: 10.1146/annurev-cellbio-101011-155807 pmid: 22804579 |
| [2] | Sears ER. Nullisomic-tetrasomic combinations in hexaploid wheat[M]//Riley R, Lewis KR. Chromosome manipulations and plant genetics. Boston: Springer, 1966:29-45. |
| [3] | Zhu B, Shao YJ, Pan Q, et al. Genome-wide gene expression perturbation induced by loss of C2 chromosome in allotetraploid Brassica napus L[J]. Front Plant Sci, 2015, 6:763. |
| [4] |
Wang M, Wang SB, Liang Z, et al. From genetic stock to genome editing:gene exploitation in wheat[J]. Trends Biotechnol, 2018, 36(2):160-172.
doi: 10.1016/j.tibtech.2017.10.002 URL |
| [5] |
Francki MG, Hayton S, Gummer JPA, et al. Metabolomic profiling and genomic analysis of wheat aneuploid lines to identify genes controlling biochemical pathways in mature grain[J]. Plant Biotechnol J, 2016, 14(2):649-660.
doi: 10.1111/pbi.12410 pmid: 26032167 |
| [6] |
Liu M, Rathjen T, Weligama K, et al. Analysis of aneuploid lines of bread wheat to map chromosomal locations of genes controlling root hair length[J]. Ann Bot, 2017, 119(8):1333-1341.
doi: 10.1093/aob/mcx030 URL |
| [7] |
Li TT, Sun YL, Liu TX, et al. TaCER1-1A is involved in cuticular wax alkane biosynthesis in hexaploid wheat and responds to plant abiotic stresses[J]. Plant Cell Environ, 2019, 42(11):3077-3091.
doi: 10.1111/pce.v42.11 URL |
| [8] |
Draeger T, C Martin A, Alabdullah AK, et al. Dmc1 is a candidate for temperature tolerance during wheat meiosis[J]. Theor Appl Genet, 2020, 133(3):809-828.
doi: 10.1007/s00122-019-03508-9 pmid: 31853574 |
| [9] | Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time[J]. Nat Rev Mol Cell Biol, 2007, 8(5):379-393. |
| [10] |
Tu YQ, Sun J, Ge XH, et al. Production and genetic analysis of partial hybrids from intertribal sexual crosses between Brassica napus and Isatis indigotica and progenies[J]. Genome, 2010, 53(2):146-156.
doi: 10.1139/g09-093 pmid: 20140033 |
| [11] |
Zhu B, Tu YQ, Zeng P, et al. Extraction of the constituent subgenomes of the natural allopolyploid rapeseed(Brassica napus L.)[J]. Genetics, 2016, 204(3):1015-1027.
doi: 10.1534/genetics.116.190967 URL |
| [12] |
Chalhoub B, Denoeud F, Liu SY, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome[J]. Science, 2014, 345(6199):950-953.
doi: 10.1126/science.1253435 pmid: 25146293 |
| [13] |
Xiong ZY, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus[J]. Proc Natl Acad Sci USA, 2011, 108(19):7908-7913.
doi: 10.1073/pnas.1014138108 URL |
| [14] |
Li Z, Liu HL, Luo P. Production and cytogenetics of intergeneric hybrids between Brassica napus and Orychophragmus violaceus[J]. Theor Appl Genet, 1995, 91(1):131-136.
doi: 10.1007/BF00220869 pmid: 24169678 |
| [15] |
Zhong XB, Jong JH, Zabel P. Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization(FISH)[J]. Chromosome Res, 1996, 4(1):24-28.
doi: 10.1007/BF02254940 pmid: 8653264 |
| [16] |
Cui C, Ge XH, Gautam M, et al. Cytoplasmic and genomic effects on meiotic pairing in Brassica hybrids and allotetraploids from pair crosses of three cultivated diploids[J]. Genetics, 2012, 191(3):725-738.
doi: 10.1534/genetics.112.140780 URL |
| [17] | Nayidu NK, Bonham-Smith P, Gruber M. Brassica villosa a potential tool to improve the insect or disease resistance of Brassica crop species[J]. Transcriptomics, 2015, 3(2):114. |
| [18] |
Alahakoon UI, Taheri A, Nayidu NK, et al. Hairy Canola(Brasssica napus)re-visited:down-regulating TTG1 in an AtGL3-enhanced hairy leaf background improves growth, leaf trichome coverage, and metabolite gene expression diversity[J]. BMC Plant Biol, 2016, 16:12.
doi: 10.1186/s12870-015-0680-5 pmid: 26739276 |
| [19] |
Nayidu NK, Kagale S, Taheri A, et al. Comparison of five major trichome regulatory genes in Brassica villosa with orthologues within the Brassicaceae[J]. PLoS One, 2014, 9(4):e95877.
doi: 10.1371/journal.pone.0095877 URL |
| [20] |
Zheng KJ, Tian HN, Hu QN, et al. Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation[J]. Sci Rep, 2016, 6:19254.
doi: 10.1038/srep19254 |
| [21] |
Li F, Chen BY, Xu K, et al. A genome-wide association study of plant height and primary branch number in rapeseed(Brassica napus)[J]. Plant Sci, 2016, 242:169-177.
doi: 10.1016/j.plantsci.2015.05.012 URL |
| [22] | Sun CM, Wang BQ, Yan L, et al. Genome-wide association study provides insight into the genetic control of plant height in rapeseed(Brassica napus L.)[J]. Front Plant Sci, 2016, 7:1102. |
| [23] |
Zheng M, Peng C, Liu HF, et al. Genome-wide association study reveals candidate genes for control of plant height, branch initiation height and branch number in rapeseed(Brassica napus L.)[J]. Front Plant Sci, 2017, 8:1246.
doi: 10.3389/fpls.2017.01246 pmid: 28769955 |
| [24] |
Li Z, McKibben MTW, Finch GS, et al. Patterns and processes of diploidization in land plants[J]. Annu Rev Plant Biol, 2021, 72:387-410.
doi: 10.1146/annurev-arplant-050718-100344 pmid: 33684297 |
| [25] |
Griffiths S, Sharp R, Foote TN, et al. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat[J]. Nature, 2006, 439(7077):749-752.
doi: 10.1038/nature04434 |
| [26] |
Serra H, Svačina R, Baumann U, et al. Ph2 encodes the mismatch repair protein MSH7-3D that inhibits wheat homoeologous recombination[J]. Nat Commun, 2021, 12(1):803.
doi: 10.1038/s41467-021-21127-1 |
| [27] |
Higgins EE, Howell EC, Armstrong SJ, et al. A major quantitative trait locus on chromosome A9, BnaPh1, controls homoeologous recombination in Brassica napus[J]. New Phytol, 2021, 229(6):3281-3293.
doi: 10.1111/nph.16986 pmid: 33020949 |
| [1] | LI Xin-yi, JIANG Chun-xiu, XUE Li, JIANG Hong-tao, YAO Wei, DENG Zu-hu, ZHANG Mu-qing, YU Fan. Enhancing Hybridization Signal of Sugarcane Chromosome Oligonucleotide Probe via Multiple Fluorescence Labeled Primers [J]. Biotechnology Bulletin, 2023, 39(5): 103-111. |
| [2] | ZHI Tian-tian, ZHOU Zhou, CHEN Ji-peng, HAN Cheng-yun. Cloning, Functional Identification and Expression Analysis of FAH, a Key Gene for Tyrosine Metabolism in Brassica napus L. [J]. Biotechnology Bulletin, 2023, 39(10): 115-127. |
| [3] | LIN Ke-yun, DUAN Yu-jing, WANG Gao-sheng, SUN Nian-li, FANG Yu-jie, WANG You-ping. Cloning and Functional Identification of BnNF-YA1 in Brassica napus L. [J]. Biotechnology Bulletin, 2022, 38(4): 106-116. |
| [4] | JIN Jiao-jiao, LIU Zi-gang, MI Wen-bo, XU Ming-xia, ZOU Ya, XU Chun-mei, ZHAO Cai-xia. Identification of Low Temperature Stress-responsive Genes Regulating Photosynthetic Characteristics in the Leaves of Brassica napus by RNA-Seq [J]. Biotechnology Bulletin, 2022, 38(4): 126-142. |
| [5] | Olalekan Amoo, HU Li-min, ZHAI Yun-gu, FAN Chu-chuan, ZHOU Yong-ming. Regulation of Shoot Branching by BRANCHED1 in Brassica napus Based on Gene Editing Technology [J]. Biotechnology Bulletin, 2022, 38(4): 97-105. |
| [6] | ZHENG Hui-qing, GUO Zhong-jie, CAI Zhi-xin, LU Yuan-ping, LIAO Jian-hua, CHEN Mei-yuan. Analysis and Evaluation of Nutrient Components in Agaricus bisporus Wild Germplasm Resource [J]. Biotechnology Bulletin, 2021, 37(11): 109-118. |
| [7] | WANG Yan-li, YANG Yi-ming, FAN Shu-tian, ZHAO Ying, XU Pei-lei, LU Wen-peng, LI Chang-yu. Genetic Diversity Analysis of 73 Vitis amurensis and Its Hybrids Offsprings Based on SSR Molecular Markers [J]. Biotechnology Bulletin, 2021, 37(1): 189-197. |
| [8] | SUN Jia-dong, SUN Xiao-feng, LI Lan, SHEN Wei, CHENG Shun-feng. Application Prospects of Stem Cell Technology in the Protection of Indigenous Porcine Germplasm Resources [J]. Biotechnology Bulletin, 2020, 36(8): 228-234. |
| [9] | SHI Jian-bin, ZHOU Hong, WANG Ning, XU Qing-hua, QIAO Wen-qing, YAN Gen-tu. Purity Identification and Genetic Diversity Analysis of Cotton Germplasm Resources Using SSR Markers [J]. Biotechnology Bulletin, 2018, 34(7): 138-146. |
| [10] | Zhang Qingtian, Fan Shutian, Yang Yiming, Li Xiaoyan, Song Rungang, Lu Wenpeng. Molecular Biology Research of Amur Grape(Vitis amurensis Rupr.) [J]. Biotechnology Bulletin, 2013, 0(12): 1-5. |
| [11] | Xu Chunbo, Wang Yong, Zhao Laixi, Zhao Haixia. Research Progress of Molecular Markers for Forage Germplasm Resources Important Traits in China [J]. Biotechnology Bulletin, 2013, 0(10): 18-23. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||