








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (4): 124-135.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0996
Previous Articles Next Articles
MA Yu-qian1( ), SUN Dong-hui1, YUE Hao-feng1, XIN Jia-yu2, LIU Ning2(
), SUN Dong-hui1, YUE Hao-feng1, XIN Jia-yu2, LIU Ning2( ), CAO Zhi-yan2,3(
), CAO Zhi-yan2,3( )
)
Received:2022-08-16
Online:2023-04-26
Published:2023-05-16
MA Yu-qian, SUN Dong-hui, YUE Hao-feng, XIN Jia-yu, LIU Ning, CAO Zhi-yan. Identification, Heterologous Expression and Functional Analysis of a GH61 Family Glycoside Hydrolase from Setosphaeria turcica with the Assisting Function in Degrading Cellulose[J]. Biotechnology Bulletin, 2023, 39(4): 124-135.
| 引物名称Primer name | 序列Sequence(5'-3') | 引物名称Primer name | 序列Sequence(5'-3') | |
|---|---|---|---|---|
| StGH61_1-F | GGCTCTGGAACTGGCAAGAT | StGH61_12-F | CTCCTGGTCCGTCACTGTTC | |
| StGH61_1-R | TAGTACTCGAGGTCCCTGGC | StGH61_12-R | GGCTGGAGAGATCGGTCATG | |
| StGH61_2-F | GTCAGTTTCCCGGGTGCTTA | StGH61_13-F | GGCAGAAGGACCAGATCGAG | |
| StGH61_2-R | GACCACGCTGTTAGTCCCAA | StGH61_13-R | CGCTGTAGAGACCGGGAATC | |
| StGH61_3-F | ACATCAACACGTGGGATCCC | StGH61_14-F | CGACGCCATCCTAGACACTC | |
| StGH61_3-R | TGCGCACGACAGGTAGAATT | StGH61_14-R | TTGAAGTCGATGCCTGGGTC | |
| StGH61_4-F | CGACTTCCGTTGCAACAAGG | StGH61_15-F | CACCAAAGTCGAGCCCTTCT | |
| StGH61_4-R | AACATCACCAGGAGCCTTGG | StGH61_15-R | TTGAGGGAGAGGGAGACGAG | |
| StGH61_5-F | TTGCGCAAATCACAACCTCG | StGH61_16-F | GTGCCAAGGGAGGTCTCTTC | |
| StGH61_5-R | TCGTGGCATGTCATGTTGGA | StGH61_16-R | GGTCGGCAGTGAAAGAGACA | |
| StGH61_6-F | CACCCAGACTGTCACCATCC | StGH61_17-F | GGTTCAAGGTGCAGGAGGAA | |
| StGH61_6-R | CACCCAGACTGTCACCATCC | StGH61_17-R | CTACCCGTCACCTTGAGCTG | |
| StGH61_7-F | GGTCCAAGTTCTCGCAGGAA | StGH61_18-F | AGCTCGACTGCCATGATCTG | |
| StGH61_7-R | CATTCACATTCGAGCCGCTG | StGH61_18-R | GGGGAGCTTGACGTTGATGA | |
| StGH61_8-F | CCACAAGAATGCTAGCCCCA | StGH61_19-F | CAAGGTCTCCAACGCAGCTA | |
| StGH61_8-R | GCACCCTCAATCTTGGTCCA | StGH61_19-R | AAAATTGAGCACCGCCAACC | |
| StGH61_9-F | TGGTTCAAGGTTTCCAGCGA | StGH61_20-F | CATCCCTGCTTGTATTGCGC | |
| StGH61_9-R | AAATGTAGAATTGCGCGCCG | StGH61_20-R | GGAACGAGACGGTTGATGGT | |
| StGH61_10-F | GGGCAGTGATGTGAAGAGCT | StGH61_21-F | AAGATCGACGAGCAAGGCAT | |
| StGH61_10-R | GGAGCACGCGAGGTAGAATT | StGH61_21-R | GACGCATACCCAGGAATCGT | |
| StGH61_11-F | AGCCTCTCTTCCTCGGACAT | β-tubulin-F | GTGCGCAAGGAGGCTGAGGG | |
| StGH61_11-R | TTTTGGCGTCGCTGACTTTG | β-tubulin-R | CATGAAGAAATGGAGACGGGGGAA |
Table 1 Primer sequences used in RT-qPCR
| 引物名称Primer name | 序列Sequence(5'-3') | 引物名称Primer name | 序列Sequence(5'-3') | |
|---|---|---|---|---|
| StGH61_1-F | GGCTCTGGAACTGGCAAGAT | StGH61_12-F | CTCCTGGTCCGTCACTGTTC | |
| StGH61_1-R | TAGTACTCGAGGTCCCTGGC | StGH61_12-R | GGCTGGAGAGATCGGTCATG | |
| StGH61_2-F | GTCAGTTTCCCGGGTGCTTA | StGH61_13-F | GGCAGAAGGACCAGATCGAG | |
| StGH61_2-R | GACCACGCTGTTAGTCCCAA | StGH61_13-R | CGCTGTAGAGACCGGGAATC | |
| StGH61_3-F | ACATCAACACGTGGGATCCC | StGH61_14-F | CGACGCCATCCTAGACACTC | |
| StGH61_3-R | TGCGCACGACAGGTAGAATT | StGH61_14-R | TTGAAGTCGATGCCTGGGTC | |
| StGH61_4-F | CGACTTCCGTTGCAACAAGG | StGH61_15-F | CACCAAAGTCGAGCCCTTCT | |
| StGH61_4-R | AACATCACCAGGAGCCTTGG | StGH61_15-R | TTGAGGGAGAGGGAGACGAG | |
| StGH61_5-F | TTGCGCAAATCACAACCTCG | StGH61_16-F | GTGCCAAGGGAGGTCTCTTC | |
| StGH61_5-R | TCGTGGCATGTCATGTTGGA | StGH61_16-R | GGTCGGCAGTGAAAGAGACA | |
| StGH61_6-F | CACCCAGACTGTCACCATCC | StGH61_17-F | GGTTCAAGGTGCAGGAGGAA | |
| StGH61_6-R | CACCCAGACTGTCACCATCC | StGH61_17-R | CTACCCGTCACCTTGAGCTG | |
| StGH61_7-F | GGTCCAAGTTCTCGCAGGAA | StGH61_18-F | AGCTCGACTGCCATGATCTG | |
| StGH61_7-R | CATTCACATTCGAGCCGCTG | StGH61_18-R | GGGGAGCTTGACGTTGATGA | |
| StGH61_8-F | CCACAAGAATGCTAGCCCCA | StGH61_19-F | CAAGGTCTCCAACGCAGCTA | |
| StGH61_8-R | GCACCCTCAATCTTGGTCCA | StGH61_19-R | AAAATTGAGCACCGCCAACC | |
| StGH61_9-F | TGGTTCAAGGTTTCCAGCGA | StGH61_20-F | CATCCCTGCTTGTATTGCGC | |
| StGH61_9-R | AAATGTAGAATTGCGCGCCG | StGH61_20-R | GGAACGAGACGGTTGATGGT | |
| StGH61_10-F | GGGCAGTGATGTGAAGAGCT | StGH61_21-F | AAGATCGACGAGCAAGGCAT | |
| StGH61_10-R | GGAGCACGCGAGGTAGAATT | StGH61_21-R | GACGCATACCCAGGAATCGT | |
| StGH61_11-F | AGCCTCTCTTCCTCGGACAT | β-tubulin-F | GTGCGCAAGGAGGCTGAGGG | |
| StGH61_11-R | TTTTGGCGTCGCTGACTTTG | β-tubulin-R | CATGAAGAAATGGAGACGGGGGAA |
| 基因 Gene | 检索号 Accession number | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 信号肽长度 Signal peptide length/aa | 蛋白定位 Protein location |
|---|---|---|---|---|---|---|
| StGH61-1 | XP_008027730 | 250 | 26.69 | 8.26 | 16 | 胞外Extracellular |
| StGH61-2 | XP_008031219 | 334 | 36.13 | 5.77 | 18 | 胞外Extracellular |
| StGH61-3 | XP_008024527 | 221 | 23.46 | 8.33 | 17 | 胞外Extracellular |
| StGH61-4 | XP_008025833 | 418 | 43.38 | 7.63 | 17 | 胞外Extracellular |
| StGH61-5 | XP_008021808 | 266 | 28.81 | 8.24 | 21 | 胞外Extracellular |
| StGH61-6 | XP_008023212 | 291 | 32.06 | 6.16 | 22 | 膜结合溶酶体Lysosomes |
| StGH61-7 | XP_008024957 | 248 | 26.32 | 7.69 | 20 | 胞外Extracellular |
| StGH61-8 | XP_008025253 | 329 | 34.56 | 5.98 | 22 | 分泌到胞外 Extracellular(Secreted) |
| StGH61-9 | XP_008030985 | 149 | 15.76 | 7.83 | — | 分泌到胞外 Extracellular(Secreted) |
| StGH61-10 | XP_008022828 | 236 | 25.55 | 9.00 | 20 | 膜结合线粒体Mitochondria |
| StGH61-11 | XP_008023697 | 229 | 23.52 | 8.49 | 16 | 胞外Extracellular |
| StGH61-12 | XP_008023696 | 258 | 27.49 | 5.47 | 22 | 胞外Extracellular |
| StGH61-13 | XP_008028494 | 325 | 34.02 | 6.31 | 18 | 胞外Extracellular |
| StGH61-14 | XP_008028319 | 292 | 30.73 | 6.28 | 21 | 胞外Extracellular |
| StGH61-15 | XP_008025489 | 472 | 49.64 | 7.44 | 19 | 胞外Extracellular |
| StGH61-16 | XP_008027303 | 222 | 23.26 | 7.69 | 20 | 胞外Extracellular |
| StGH61-17 | XP_008027295 | 246 | 25.92 | 8.44 | 18 | 胞外Extracellular |
| StGH61-18 | XP_008024355 | 344 | 34.75 | 6.49 | 17 | 胞外Extracellular |
| StGH61-19 | XP_008022590 | 243 | 25.06 | 8.90 | 17 | 胞外Extracellular |
| StGH61-20 | XP_008026441 | 233 | 24.21 | 7.67 | 18 | 胞外Extracellular |
Table 2 Prediction of the protein properties of the glycoside hydrolase GH61 family from S. turcica
| 基因 Gene | 检索号 Accession number | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 信号肽长度 Signal peptide length/aa | 蛋白定位 Protein location |
|---|---|---|---|---|---|---|
| StGH61-1 | XP_008027730 | 250 | 26.69 | 8.26 | 16 | 胞外Extracellular |
| StGH61-2 | XP_008031219 | 334 | 36.13 | 5.77 | 18 | 胞外Extracellular |
| StGH61-3 | XP_008024527 | 221 | 23.46 | 8.33 | 17 | 胞外Extracellular |
| StGH61-4 | XP_008025833 | 418 | 43.38 | 7.63 | 17 | 胞外Extracellular |
| StGH61-5 | XP_008021808 | 266 | 28.81 | 8.24 | 21 | 胞外Extracellular |
| StGH61-6 | XP_008023212 | 291 | 32.06 | 6.16 | 22 | 膜结合溶酶体Lysosomes |
| StGH61-7 | XP_008024957 | 248 | 26.32 | 7.69 | 20 | 胞外Extracellular |
| StGH61-8 | XP_008025253 | 329 | 34.56 | 5.98 | 22 | 分泌到胞外 Extracellular(Secreted) |
| StGH61-9 | XP_008030985 | 149 | 15.76 | 7.83 | — | 分泌到胞外 Extracellular(Secreted) |
| StGH61-10 | XP_008022828 | 236 | 25.55 | 9.00 | 20 | 膜结合线粒体Mitochondria |
| StGH61-11 | XP_008023697 | 229 | 23.52 | 8.49 | 16 | 胞外Extracellular |
| StGH61-12 | XP_008023696 | 258 | 27.49 | 5.47 | 22 | 胞外Extracellular |
| StGH61-13 | XP_008028494 | 325 | 34.02 | 6.31 | 18 | 胞外Extracellular |
| StGH61-14 | XP_008028319 | 292 | 30.73 | 6.28 | 21 | 胞外Extracellular |
| StGH61-15 | XP_008025489 | 472 | 49.64 | 7.44 | 19 | 胞外Extracellular |
| StGH61-16 | XP_008027303 | 222 | 23.26 | 7.69 | 20 | 胞外Extracellular |
| StGH61-17 | XP_008027295 | 246 | 25.92 | 8.44 | 18 | 胞外Extracellular |
| StGH61-18 | XP_008024355 | 344 | 34.75 | 6.49 | 17 | 胞外Extracellular |
| StGH61-19 | XP_008022590 | 243 | 25.06 | 8.90 | 17 | 胞外Extracellular |
| StGH61-20 | XP_008026441 | 233 | 24.21 | 7.67 | 18 | 胞外Extracellular |
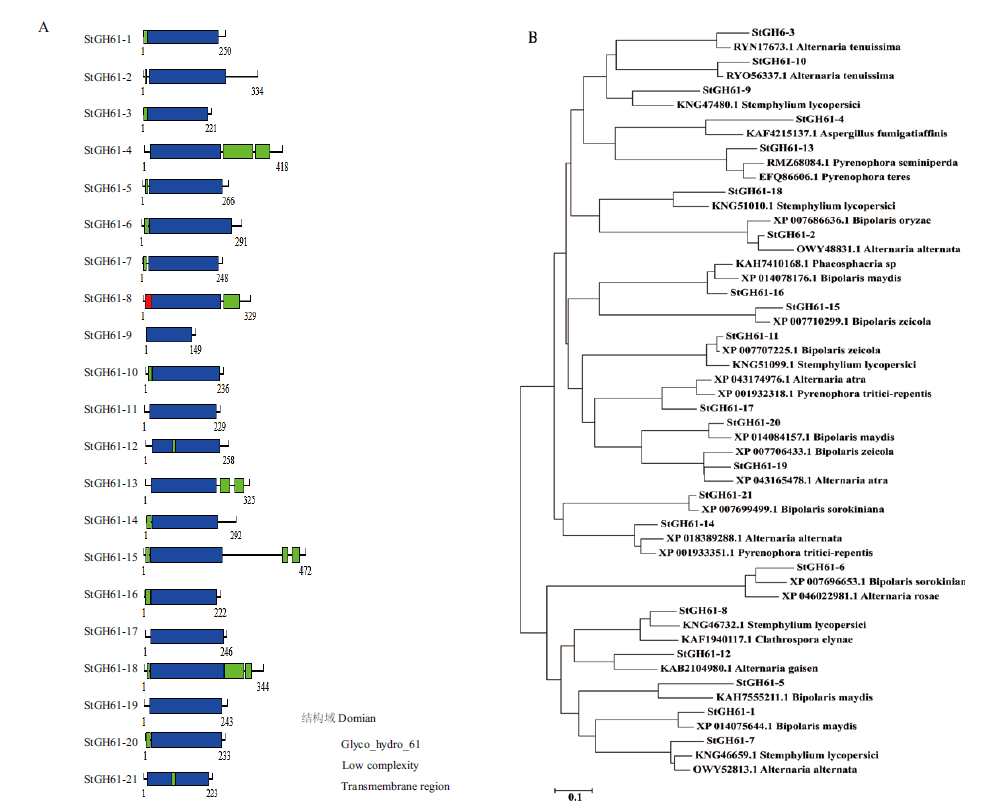
Fig. 1 Protein domains and evolutionary analysis of the GH61 family of glycoside hydrolases from S. turcica A: Domain of GH61 family of glycoside hydrolases from S. turcica. B: Evolutionary relationship of glycoside hydrolases GH61 family proteins from S. turcica
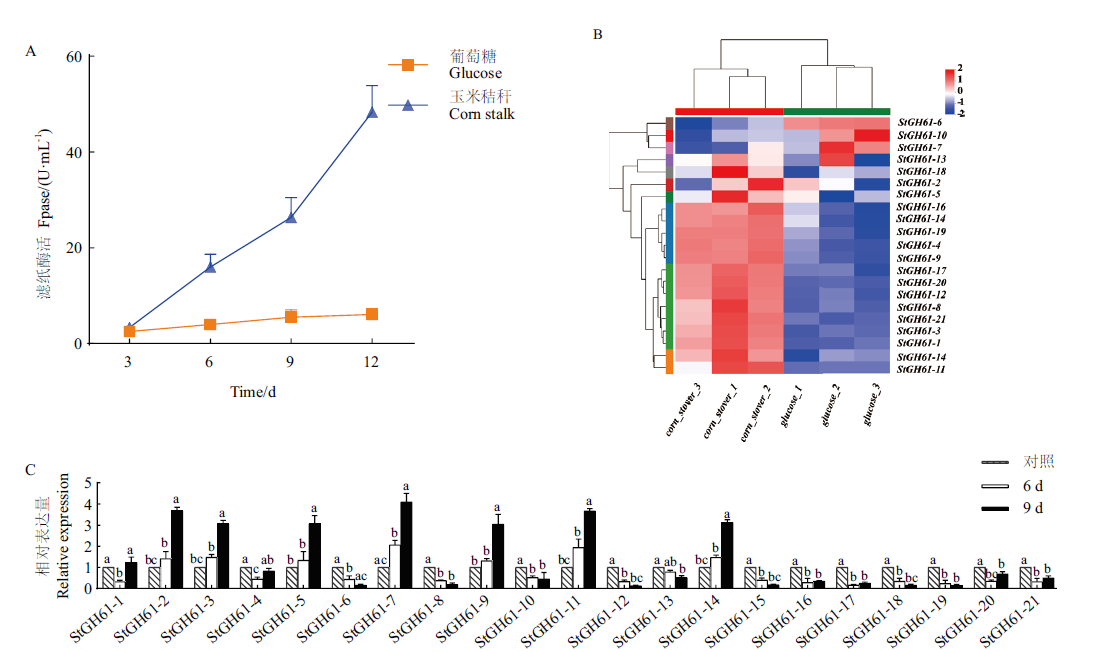
Fig. 2 Filter paper enzymatic activity and GH61 family gene expression analysis of cellulase from S. turcica A: Filter paper enzymatic activity of cellulase from S. turcica. B: Expression map of GH61 family glycoside hydrolase genes in S. turcica. C: RT-qPCR verifies the relative expression of GH61 family glycoside hydrolase genes in S. turcica. Different normal letters indicate significant differences among different treatments at 0.05 level

Fig. 3 Heterologous expression of StGH61-11 recombinant protein and optimized induction conditions A: Agarose gel electrophoresis results of the target gene StGH61-11 PCR products, 1-3: GH61-11 cDNA fragment. M:DNA marker. B: Agarose gel electrophoresis results of PCR for recombinant transformants, 1-3: Pet32a-StGH61-11 recombinant plasmid, M:DNA marker. C: SDS-PAGE detection of whole bacterial lysate expressed by recombinant pET32a-StGH61-11(BL21), 0 : Pet32a-StGH61-11-BL21(DE3)not induce control, 1 : Pet32a-StGH61-11-BL21(DE3)the recombinant bacteria were induced by IPTG, M : Standard for molecular weight of pre-dyed protein. D: Effect of induction time on the enzymatic activity of recombinant protein StGH61-11. E: Effect of IPTG concentration on the enzymatic activity of recombinant protein StGH61-11
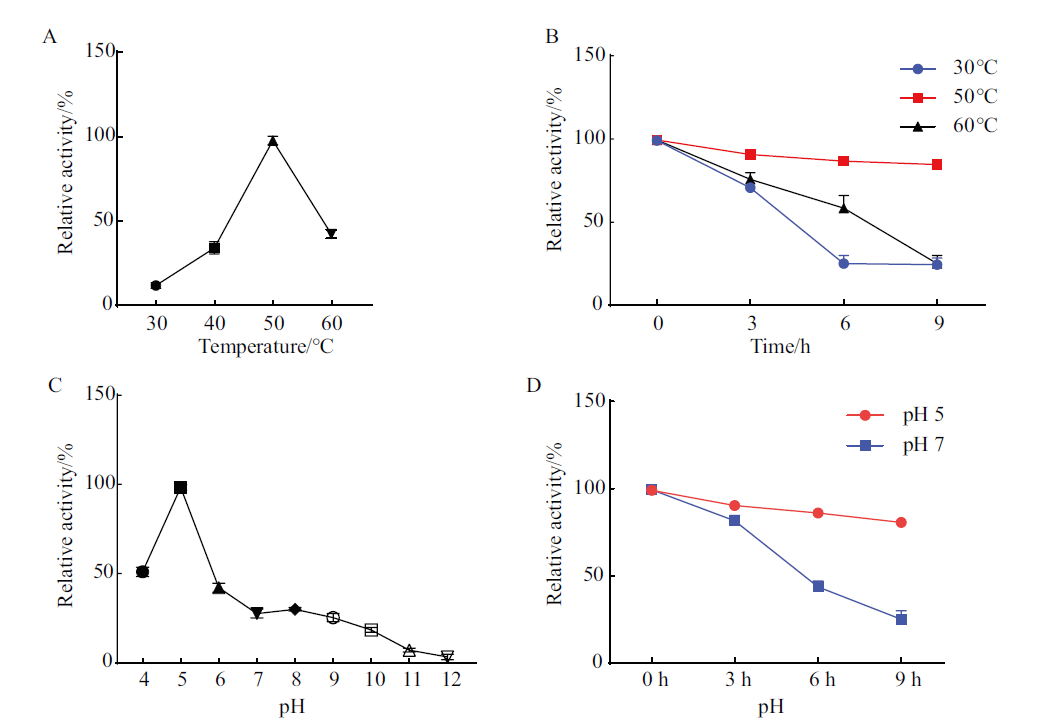
Fig. 4 Optimal reaction temperature and stability and pH and stability of StGH61-11 recombinant protein A: Effects of temperature on the cellulase activity of StGH61-11. B: Effect of temperature on the stability of StGH61-11 cellulase activity. C: Effects of pH on StGH61-11 cellulase activity. D: Effect of pH on the viability stability of StGH61-11 cellulase
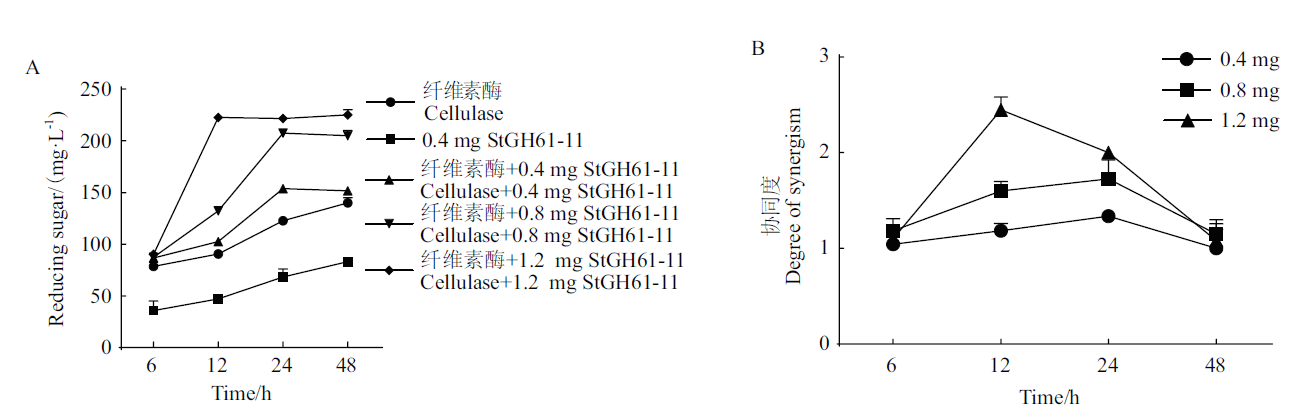
Fig. 5 Synergistic reaction of StGH61-11 with cellulase hydrolysis of corn stover A: Promoting effect of StGH61-11 on cellulase activity. B: Synergism degree of StGH61-11 and cellulase
| [1] |
Huang Z, Ni GR, Zhao XY, et al. Characterization of a GH8 β-1, 4-glucanase from Bacillus subtilis B111 and its saccharification potential for agricultural straws[J]. J Microbiol Biotechnol, 2021, 31(10): 1446-1454.
doi: 10.4014/jmb.2105.05026 URL |
| [2] | 许从峰, 艾士奇, 申贵男, 等. 木质纤维素的微生物降解[J]. 生物工程学报, 2019, 35(11): 2081-2091. |
| Xu CF, Ai SQ, Shen GN, et al. Microbial degradation of lignocellulose[J]. Chin J Biotechnol, 2019, 35(11): 2081-2091. | |
| [3] |
Østby H, Hansen LD, Horn SJ, et al. Enzymatic processing of lignocellulosic biomass: principles, recent advances and perspectives[J]. J Ind Microbiol Biotechnol, 2020, 47(9/10): 623-657.
doi: 10.1007/s10295-020-02301-8 URL |
| [4] |
Zhang WR, Wang WW, Wang JH, et al. Isolation and characterization of a novel laccase for lignin degradation, LacZ1[J]. Appl Environ Microbiol, 2021, 87(23): e0135521.
doi: 10.1128/AEM.01355-21 URL |
| [5] |
Kucharska K, Rybarczyk P, Hołowacz I, et al. Pretreatment of lignocellulosic materials as substrates for fermentation processes[J]. Molecules, 2018, 23(11): 2937.
doi: 10.3390/molecules23112937 URL |
| [6] |
Wang YS, Shao Y, Zou XY, et al. Synergistic action between extracellular products from white-rot fungus and cellulase significantly improves enzymatic hydrolysis[J]. Bioengineered, 2018, 9(1): 178-185.
doi: 10.1080/21655979.2017.1308991 pmid: 28384075 |
| [7] | 饶佳, 鲍大鹏, 李燕, 等. 草菇GH61家族基因的生物信息学分析及金属离子对其表达水平的影响[J]. 菌物学报, 2016, 35(5): 586-596. |
| Rao J, Bao DP, Li Y, et al. Bioinformatic and gene expression analyses of the GH61 family genes of Volvariella volvacea[J]. Mycosystema, 2016, 35(5): 586-596. | |
| [8] |
Sun PC, Valenzuela SV, Chunkrua P, et al. Oxidized product profiles of AA9 lytic polysaccharide monooxygenases depend on the type of cellulose[J]. ACS Sustain Chem Eng, 2021, 9(42): 14124-14133.
doi: 10.1021/acssuschemeng.1c04100 pmid: 34722005 |
| [9] |
Waghmare PR, Waghmare PP, Gao LW, et al. Efficient constitutive expression of cellulolytic enzymes in Penicillium oxalicum for improved efficiency of lignocellulose degradation[J]. J Microbiol Biotechnol, 2021, 31(5): 740-746.
doi: 10.4014/jmb.2101.01003 URL |
| [10] |
Vaaje-Kolstad G, Westereng B, Horn SJ, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides[J]. Science, 2010, 330(6001): 219-222.
doi: 10.1126/science.1192231 pmid: 20929773 |
| [11] |
Morgenstern I, Powlowski J, Tsang A. Fungal cellulose degradation by oxidative enzymes: from dysfunctional GH61 family to powerful lytic polysaccharide monooxygenase family[J]. Brief Funct Genomics, 2014, 13(6): 471-481.
doi: 10.1093/bfgp/elu032 pmid: 25217478 |
| [12] |
Pierce BC, Agger JW, Zhang ZH, et al. A comparative study on the activity of fungal lytic polysaccharide monooxygenases for the depolymerization of cellulose in soybean spent flakes[J]. Carbohydr Res, 2017, 449: 85-94.
doi: 10.1016/j.carres.2017.07.004 URL |
| [13] |
Zhang RQ. Functional characterization of cellulose-degrading AA9 lytic polysaccharide monooxygenases and their potential exploitation[J]. Appl Microbiol Biotechnol, 2020, 104(8): 3229-3243.
doi: 10.1007/s00253-020-10467-5 pmid: 32076777 |
| [14] |
Mazurkewich S, Seveso A, Hüttner S, et al. Structure of a C1/C4-oxidizing AA9 lytic polysaccharide monooxygenase from the thermophilic fungus Malbranchea cinnamomea[J]. Acta Crystallogr D Struct Biol, 2021, 77(Pt 8): 1019-1026.
doi: 10.1107/S2059798321006628 URL |
| [15] |
Meng YN, Zeng FL, Hu JJ, et al. Novel factors contributing to fungal pathogenicity at early stages of Setosphaeria turcica infection[J]. Mol Plant Pathol, 2022, 23(1): 32-44.
doi: 10.1111/mpp.13140 URL |
| [16] | 王晶晶. StPP2A-c基因调控玉米大斑病菌致病性的机制研究[D]. 保定: 河北农业大学, 2013. |
| Wang JJ. The mechanism of StPP2A-c gene regulating the pathogenicity in Setosphaeria turcica[D]. Baoding: Hebei Agricultural University, 2013. | |
| [17] |
常晴, 束月蓉, 王文韬, 等. 来自Yeosuana marina sp. JLT21内切型海藻酸裂解酶的异源表达及酶学表征[J]. 生物技术通报, 2022, 38(2): 123-131.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-0539 |
| Chang Q, Shu YR, Wang WT, et al. Heterologous expression and characterization of endo-type alginate lyase from Yeosuana marina sp. JLT21[J]. Biotechnol Bull, 2022, 38(2): 123-131. | |
| [18] |
薛鲜丽, 王静然, 毕杭杭, 等. 过表达Spt7对黑曲霉生长及抗逆性影响[J]. 生物技术通报, 2022, 38(5): 112-122.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-0946 |
| Xue XL, Wang JR, Bi HH, et al. Effect of Spt7 overexpression of on the growth and stress resistance of Aspergillus niger[J]. Biotechnol Bull, 2022, 38(5): 112-122. | |
| [19] |
Breslmayr E, Hanžek M, Hanrahan A, et al. A fast and sensitive activity assay for lytic polysaccharide monooxygenase[J]. Biotechnol Biofuels, 2018, 11: 79.
doi: 10.1186/s13068-018-1063-6 pmid: 29588664 |
| [20] |
de Gouvêa PF, Bernardi AV, Gerolamo LE, et al. Transcriptome and secretome analysis of Aspergillus fumigatus in the presence of sugarcane bagasse[J]. BMC Genomics, 2018, 19(1): 232.
doi: 10.1186/s12864-018-4627-8 |
| [21] | 白雪, 张梦娣, 张东远, 等. 丝状真菌溶解性多糖单加氧酶的研究进展[J]. 基因组学与应用生物学, 2018, 37(12): 5339-5348. |
| Bai X, Zhang MD, Zhang DY, et al. Research progress on lytic polysaccharide monooxygenases(LPMOs)in filamentous fungi[J]. Genom Appl Biol, 2018, 37(12): 5339-5348. | |
| [22] |
Midorikawa GEO, Correa CL, Noronha EF, et al. Analysis of the transcriptome in Aspergillus tamarii during enzymatic degradation of sugarcane bagasse[J]. Front Bioeng Biotechnol, 2018, 6: 123.
doi: 10.3389/fbioe.2018.00123 URL |
| [23] |
de Gouvêa PF, Gerolamo LE, Bernardi AV, et al. Lytic polysaccharide monooxygenase from Aspergillus fumigatus can improve enzymatic cocktail activity during sugarcane bagasse hydrolysis[J]. Protein Pept Lett, 2019, 26(5): 377-385.
doi: 10.2174/0929866526666190228163629 URL |
| [24] | 郭宵, 安亚静, 柴成程, 等. 大肠杆菌分泌表达裂解性多糖单加氧酶发酵条件的优化[J]. 食品与发酵工业, 2020, 46(5): 31-37. |
| Guo X, An YJ, Chai CC, et al. Fermentation condition optimization of recombinant lytic polysaccharide monooxygenase extracellularly expressed in Escherichia coli[J]. Food Ferment Ind, 2020, 46(5): 31-37. | |
| [25] |
Bernardi AV, Gerolamo LE, de Gouvêa PF, et al. LPMO af AA9_B and cellobiohydrolase af Cel6A from A. fumigatus boost enzymatic saccharification activity of cellulase cocktail[J]. Int J Mol Sci, 2020, 22(1): 276.
doi: 10.3390/ijms22010276 URL |
| [26] | 冯玉和, 孙小宝, 陈书昕, 等. 米曲霉裂解性多糖单加氧酶的异源表达与性质分析[J]. 微生物学报, 2020, 60(1): 183-199. |
| Feng YH, Sun XB, Chen SX, et al. Heterologous expression and characterization of Aspergillus oryzae lytic polysaccharide monooxygenases[J]. Acta Microbiol Sin, 2020, 60(1): 183-199. | |
| [27] | 夏东慧, 刘宁, 郭秀娜, 等. 嗜热毛壳菌多糖单加氧酶的氧化特性及协同作用[J]. 菌物学报, 2022, 41(7): 1068-1079. |
| Xia DH, Liu N, Guo XN, et al. The oxidation properties and synergism of polysaccharide monooxygenase from Chaetomium thermophilum[J]. Mycosystema, 2022, 41(7): 1068-1079. | |
| [28] |
Singh RK, Blossom BM, Russo DA, et al. Thermal unfolding and refolding of a lytic polysaccharide monooxygenase from Thermoascus aurantiacus[J]. RSC Adv, 2019, 9(51): 29734-29742.
doi: 10.1039/C9RA05920B URL |
| [29] |
Tuveng TR, Jensen MS, Fredriksen L, et al. A thermostable bacterial lytic polysaccharide monooxygenase with high operational stability in a wide temperature range[J]. Biotechnol Biofuels, 2020, 13(1): 194.
doi: 10.1186/s13068-020-01834-5 pmid: 33292445 |
| [30] | 马清. 黑曲霉多糖单加氧酶的克隆表达与协同性研究[D]. 天津: 天津科技大学, 2018. |
| Ma Q. Cloning of lytic polysaccharide monooxygenases genes from asperillus Niger and research on its synergism activity[D]. Tianjin: Tianjin University of Science & Technology, 2018. |
| [1] | RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability [J]. Biotechnology Bulletin, 2023, 39(8): 283-290. |
| [2] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [3] | ZHANG Jing, ZHANG Hao-rui, CAO Yun, HUANG Hong-ying, QU Ping, ZHANG Zhi-ping. Research Progress in Thermophilic Microorganisms for Cellulose Degradation [J]. Biotechnology Bulletin, 2023, 39(6): 73-87. |
| [4] | CHEN Nan-nan, WANG Chun-lai, JIANG Zhen-zhong, JIAO Peng, GUAN Shu-yan, MA Yi-yong. Genetic Transformation and Chilling Resistance Analysis of Maize ZmDHN15 Gene in Tobacco [J]. Biotechnology Bulletin, 2023, 39(4): 259-267. |
| [5] | ZHANG Kai-ping, LIU Yan-li, TU Mian-liang, LI Ji-wei, WU Wen-biao. Optimization of Producing Cellulase by Aspergillus fumigatus A-16 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2022, 38(9): 215-225. |
| [6] | WANG Xin-guang, TIAN Lei, WANG En-ze, ZHONG Cheng, TIAN Chun-jie. Construction of Microbial Consortium for Efficient Degradation of Corn Straw and Evaluation of Its Degradation Effect [J]. Biotechnology Bulletin, 2022, 38(4): 217-229. |
| [7] | NIU Xin, ZHANG Ying, WANG Mao-jun, LIU Wen-long, LU Fu-ping, LI Yu. Effects of Different Integration Sites on the Expression of Exogenous Alkaline Protease in Bacillus amyloliquefaciens [J]. Biotechnology Bulletin, 2022, 38(4): 253-260. |
| [8] | ZHANG Gong-you, WANG Yi-han, GUO Min, ZHANG Ting-ting, WANG Bing, LIU Hong-mei. Isolation and Identification of a Cellulase-producing Endophytic Fungus in Paris polyphylla var. yunnanensis [J]. Biotechnology Bulletin, 2022, 38(2): 95-104. |
| [9] | WANG Bo-ya, JIANG Yong, HUANG Yan, CAO Ying, HU Shang-lian. Cloning and Functional Analysis of BeCesA4 in Bambusa emeiensis [J]. Biotechnology Bulletin, 2022, 38(11): 185-193. |
| [10] | WANG Xiao-tao, ZOU Hang, WU Yi, XIANG Shen-wei, LV Hua, LIU Chao-lan, LIN Jia-fu, WANG Xin-rong, CHU Yi-wen, SONG Tao. Heterologous Expression and Enzymatic Properties Analysis of Novel β-agarase Aga2 from Paraglaciecola hydrolytica [J]. Biotechnology Bulletin, 2022, 38(11): 258-268. |
| [11] | ZHANG Tong-tong, ZHENG Deng-yu, WU Zhong-yi, ZHANG Zhong-bao, YU Rong. Functional Analysis of ZmNF-YB13 Responding to Drought and Salt Stress [J]. Biotechnology Bulletin, 2022, 38(10): 115-123. |
| [12] | TANG Hao, SUN Can, LI Yuan-qiu, LUO Chao-bing. Screening and Genome Sequencing of Cellulytic Bacterium Raoultella ornithinolytica LL1 [J]. Biotechnology Bulletin, 2021, 37(6): 85-96. |
| [13] | LIAO Zhao-min, CAI Jun, LIN Jian-guo, DU Xin, WANG Chang-gao. Expression of Glucose Oxidase Gene from Aspergillus niger in Pichia pastoris and Optimization of Enzyme Production Conditions [J]. Biotechnology Bulletin, 2021, 37(6): 97-107. |
| [14] | LIU Shan, YE Wei, ZHU Mu-zi, LI Sai-ni, DENG Zhang-shuang, ZHANG Wei-min. Cloning,Expression and Characterization of a Novel Acyltransferase GPAT [J]. Biotechnology Bulletin, 2021, 37(11): 257-266. |
| [15] | HU Fang, DONG Xu, SHI Chang-wei, WU Xue-dong. Progress in Ultrasound Intensification for Enzymatic Hydrolysis of Lignocellulose [J]. Biotechnology Bulletin, 2021, 37(10): 234-244. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||