








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (4): 288-296.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0917
Previous Articles Next Articles
YAO Jin-dong1,2,3( ), TANG Hua-mei1,2,3, YANG Wen-xiao1,2,3, ZHANG Li-shan1,2,3, LIN Xiang-min1,2,3(
), TANG Hua-mei1,2,3, YANG Wen-xiao1,2,3, ZHANG Li-shan1,2,3, LIN Xiang-min1,2,3( )
)
Received:2022-07-24
Online:2023-04-26
Published:2023-05-16
YAO Jin-dong, TANG Hua-mei, YANG Wen-xiao, ZHANG Li-shan, LIN Xiang-min. Comparative Proteomics Analysis of Aeromonas hydrophila Under Enrofloxacin Stress[J]. Biotechnology Bulletin, 2023, 39(4): 288-296.
| Gene | Oligonucleotide sequence(5'-3') | Purpose |
|---|---|---|
| cysA-F | ATCGGGTGGTGCTGATGAACG | RT-qPCR |
| cysA-R | TGGGTAGCAGGCGGGTGATG | RT-qPCR |
| cysN-F | GGAGTCCGACAGCCAGAAG | RT-qPCR |
| cysN-R | GGCGGTGGAGAAATAGCG | RT-qPCR |
| cysD-F | GCTGGACATCTGGCAATACA | RT-qPCR |
| cysD-R | GCTCCTGCTTCACTTCATCTT | RT-qPCR |
| cysI-F | TCCACGCCAACGATCTCAA | RT-qPCR |
| cysI-R | GCCACGGCTGGTAAACTCAT | RT-qPCR |
| cysH-F | CCCCATCAATCGGGCTCT | RT-qPCR |
| cysH-R | TCCATCGGCTCCACCTTG | RT-qPCR |
| cysG2-F | CGCCTTTGCCTCCAACACC | RT-qPCR |
| cysG2-R | CCTTCTTGCCGACCGACACC | RT-qPCR |
| cysC-F | TATCGTGCTCACCGCCTTTA | RT-qPCR |
| cysC-R | CCTGCCCTAGCCTTCTTGTAG | RT-qPCR |
| cysK-F | ATCGGCGCCAACCTGATCT | RT-qPCR |
| cysK-R | CATTGCCAAGGCCAACGAG | RT-qPCR |
Table 1 Primer pairs used in this study
| Gene | Oligonucleotide sequence(5'-3') | Purpose |
|---|---|---|
| cysA-F | ATCGGGTGGTGCTGATGAACG | RT-qPCR |
| cysA-R | TGGGTAGCAGGCGGGTGATG | RT-qPCR |
| cysN-F | GGAGTCCGACAGCCAGAAG | RT-qPCR |
| cysN-R | GGCGGTGGAGAAATAGCG | RT-qPCR |
| cysD-F | GCTGGACATCTGGCAATACA | RT-qPCR |
| cysD-R | GCTCCTGCTTCACTTCATCTT | RT-qPCR |
| cysI-F | TCCACGCCAACGATCTCAA | RT-qPCR |
| cysI-R | GCCACGGCTGGTAAACTCAT | RT-qPCR |
| cysH-F | CCCCATCAATCGGGCTCT | RT-qPCR |
| cysH-R | TCCATCGGCTCCACCTTG | RT-qPCR |
| cysG2-F | CGCCTTTGCCTCCAACACC | RT-qPCR |
| cysG2-R | CCTTCTTGCCGACCGACACC | RT-qPCR |
| cysC-F | TATCGTGCTCACCGCCTTTA | RT-qPCR |
| cysC-R | CCTGCCCTAGCCTTCTTGTAG | RT-qPCR |
| cysK-F | ATCGGCGCCAACCTGATCT | RT-qPCR |
| cysK-R | CATTGCCAAGGCCAACGAG | RT-qPCR |
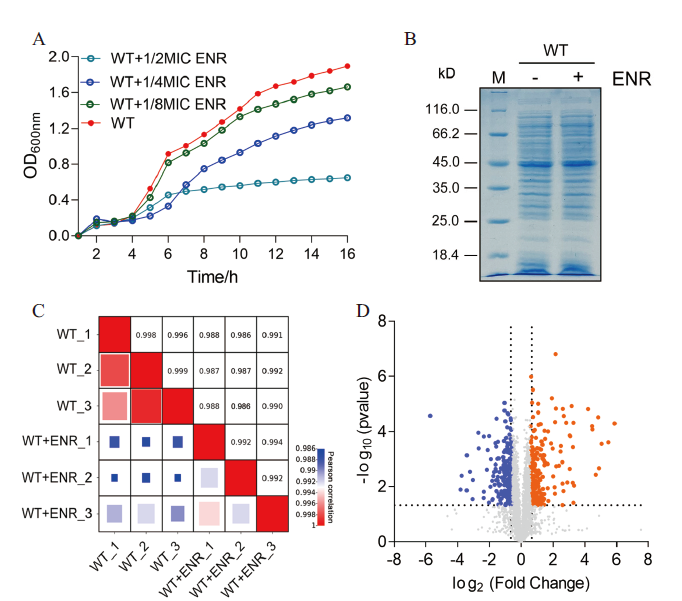
Fig. 1 Analysis of quantitative proteomics data A: Growth curves of A. hydrophila under different concentrations of enrofloxacin stress. B: SDS-PAGE profiles of A. hydrophila with or without enrofloxacin treatment. C: Correlation analysis of protein expression of three biological replicates in each group was carried out by correlation coefficient. D: Volcanic plots showing the abundance of differential protein expression. The blue dots represent the differentially downregulated proteins, the orange dots represent the differentially up-regulated proteins, and the gray dots represent the non-differentially expressed proteins

Fig. 2 GO and KEGG enrichment analysis of differentially expressed proteins under ENR stress A: Using DAVID online website and R language to analyze the biological processes of differentially expressed proteins. B: Molecular function. C: Enrichment analysis of KEGG metabolic pathway

Fig. 3 Protein-protein interaction prediction(PPI)net-work of differential proteins under ENR stress The extended network of hub proteins. Different colors indicate the fold change of different proteins(Log2)
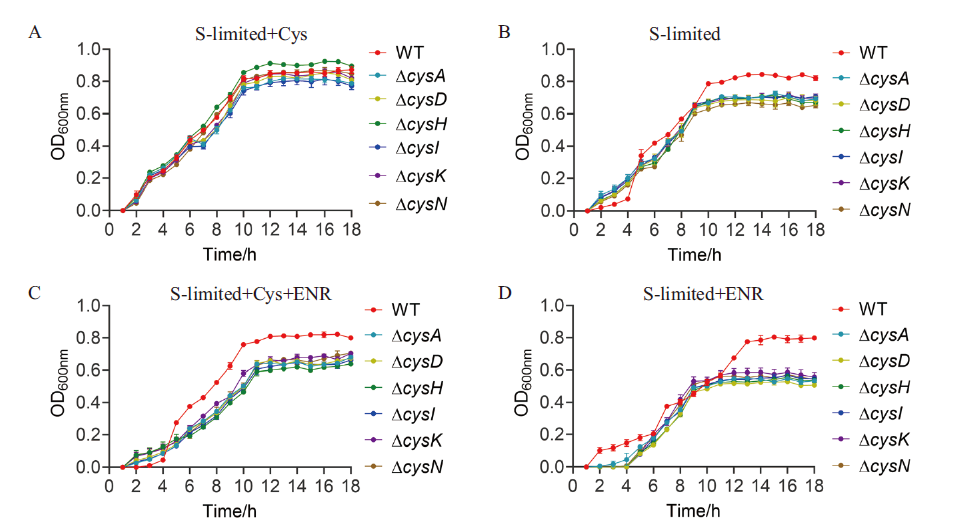
Fig. 4 Growths of WT and sulfur metabolism-related gene deletion strains supplemented with 2 mmol/L cysteine and ENR in sulfur limiting medium, respectively A: 2 mmol/L cysteine was added to the sulfur-limited medium. B: Sulfur-limited medium. C: Sulfur limited medium with 2 mmol/L cysteine and 0.78 µg/mL ENR. D: ENR was added to the sulfur-limited medium

Fig. 5 qPCR validation of proteomic data qPCR was used to verify the mRNA expression levels of 8 sulfur metabolism related genes in A. hydrophila under ENR stress or not
| [1] |
Harikrishnan R, Balasundaram C. Modern trends in Aeromonas hydrophila disease management with fish[J]. Rev Fish Sci, 2005, 13(4): 281-320.
doi: 10.1080/10641260500320845 URL |
| [2] |
Adamski J, Koivuranta M, Leppänen E. Fatal case of myonecrosis and septicaemia caused by Aeromonas hydrophila in Finland[J]. Scand J Infect Dis, 2006, 38(11-12): 1117-1119.
pmid: 17148092 |
| [3] |
Dahanayake PS, Hossain S, Wickramanayake MVKS, et al. Antibiotic and heavy metal resistance genes in Aeromonas spp. isolated from marketed Manila Clam(Ruditapes philippinarum)in Korea[J]. J Appl Microbiol, 2019, 127(3): 941-952.
doi: 10.1111/jam.14355 pmid: 31211903 |
| [4] |
Stratev D, Odeyemi OA. Antimicrobial resistance of Aeromonas hydrophila isolated from different food sources: a mini-review[J]. J Infect Public Health, 2016, 9(5): 535-544.
doi: 10.1016/j.jiph.2015.10.006 URL |
| [5] |
Paudel S, Peña-Bahamonde J, Shakiba S, et al. Prevention of infection caused by enteropathogenic E. coli O157: H7 in intestinal cells using enrofloxacin entrapped in polymer based nanocarriers[J]. J Hazard Mater, 2021, 414: 125454.
doi: 10.1016/j.jhazmat.2021.125454 URL |
| [6] |
Mitchell MA. Enrofloxacin[J]. J Exot Pet Med, 2006, 15(1): 66-69.
doi: 10.1053/j.jepm.2005.11.011 URL |
| [7] |
Sun J, Huang XY, Dong YP, et al. Transcriptome responses in enrofloxacin-resistant strains of Aeromonas hydrophila exposed to Forsythia suspensa[J]. Aquaculture, 2020, 517: 734809.
doi: 10.1016/j.aquaculture.2019.734809 URL |
| [8] |
Zhu FJ, Yang ZY, Zhang YL, et al. Transcriptome differences between enrofloxacin-resistant and enrofloxacin-susceptible strains of Aeromonas hydrophila[J]. PLoS One, 2017, 12(7): e0179549.
doi: 10.1371/journal.pone.0179549 URL |
| [9] |
Sha S, Dong ZX, Gao YS, et al. In-situ removal of residual antibiotics(enrofloxacin)in recirculating aquaculture system: effect of ultraviolet photolysis plus biodegradation using immobilized microbial granules[J]. J Clean Prod, 2022, 333: 130190.
doi: 10.1016/j.jclepro.2021.130190 URL |
| [10] |
Tanca A, Biosa G, Pagnozzi D, et al. Comparison of detergent-based sample preparation workflows for LTQ-Orbitrap analysis of the Escherichia coli proteome[J]. Proteomics, 2013, 13(17): 2597-2607.
doi: 10.1002/pmic.201200478 URL |
| [11] |
Wang GB, Wang YQ, Zhang LS, et al. Proteomics analysis reveals the effect of Aeromonas hydrophila sirtuin CobB on biological functions[J]. J Proteomics, 2020, 225: 103848.
doi: 10.1016/j.jprot.2020.103848 URL |
| [12] |
Huang DW, Sherman BT, Tan QN, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists[J]. Nucleic Acids Res, 2007, 35(Web Server issue): W169-W175.
doi: 10.1093/nar/gkm415 pmid: 17576678 |
| [13] |
Li Z, Zhang LS, Sun LN, et al. Proteomics analysis reveals the importance of transcriptional regulator slyA in regulation of several physiological functions in Aeromonas hydrophila[J]. J Proteomics, 2021, 244: 104275.
doi: 10.1016/j.jprot.2021.104275 URL |
| [14] | Yu J, Ramanathan S, Chen LC, et al. Comparative transcriptomic analysis reveals the molecular mechanisms related to oxytetracycline- resistance in strains of Aeromonas hydrophila[J]. Aquac Rep, 2021, 21: 100812. |
| [15] |
Cai QL, Wang GB, Li ZQ, et al. SWATH based quantitative proteomics analysis reveals Hfq2 play an important role on pleiotropic physiological functions in Aeromonas hydrophila[J]. J Proteomics, 2019, 195: 1-10.
doi: 10.1016/j.jprot.2018.12.030 URL |
| [16] |
Erwin S, Foster DM, Jacob ME, et al. The effect of enrofloxacin on enteric Escherichia coli: fitting a mathematical model to in vivo data[J]. PLoS One, 2020, 15(1): e0228138.
doi: 10.1371/journal.pone.0228138 URL |
| [17] |
Bush NG, Diez-Santos I, Abbott LR, et al. Quinolones: mechanism, lethality and their contributions to antibiotic resistance[J]. Molecules, 2020, 25(23): 5662.
doi: 10.3390/molecules25235662 URL |
| [18] |
Pham TDM, Ziora ZM, Blaskovich MAT. Quinolone antibiotics[J]. MedChemComm, 2019, 10(10): 1719-1739.
doi: 10.1039/c9md00120d pmid: 31803393 |
| [19] |
Li J, Wang T, Shao B, et al. Plasmid-mediated quinolone resistance genes and antibiotic residues in wastewater and soil adjacent to swine feedlots: potential transfer to agricultural lands[J]. Environ Health Perspect, 2012, 120(8): 1144-1149.
doi: 10.1289/ehp.1104776 URL |
| [20] |
Girijan SK, Paul R, V J RK, et al. Investigating the impact of hospital antibiotic usage on aquatic environment and aquaculture systems: a molecular study of quinolone resistance in Escherichia coli[J]. Sci Total Environ, 2020, 748: 141538.
doi: 10.1016/j.scitotenv.2020.141538 URL |
| [21] |
Peng B, Su YB, Li H, et al. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria[J]. Cell Metab, 2015, 21(2): 249-262.
doi: S1550-4131(15)00009-1 pmid: 25651179 |
| [1] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [2] | YAN Xiong-ying, WANG Zhen, WANG Xia, YANG Shi-hui. Microbial Sulfur Metabolism and Stress Resistance [J]. Biotechnology Bulletin, 2023, 39(11): 150-167. |
| [3] | GAO Xue-yan, CHEN Lin-xu, CHEN Xian-ke, PANG Xin, PAN Deng, LIN Jian-qun. Application of Acidithiobacillus spp. in Industry and Agriculture [J]. Biotechnology Bulletin, 2022, 38(5): 36-46. |
| [4] | Zheng Juanjuan, Zhang Yu, Xu Wentao, Huang Kunlun. Overview of the Protection Effect of Zinc on Mammalian Cell Damage [J]. Biotechnology Bulletin, 2013, 0(4): 21-26. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||