








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (5): 1-13.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1098
FENG Shan-shan( ), WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie(
), WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie( )
)
Received:2022-09-06
Online:2023-05-26
Published:2023-06-08
Contact:
FANG Yu-jie
E-mail:fss7627@163.com;yjfang@yzu.edu.cn
FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response[J]. Biotechnology Bulletin, 2023, 39(5): 1-13.

Fig. 1 Phylogenetic evolution tree of WOX family The phylogenetic tree was constructed by MEGA7 software using the amino acid sequences of WOX members in Physcomitrella patens,Selaginella moellendorffii,Picea abies,Amborella trichopoda,Oryza sativa,and Arabidopsis thaliana WOX family by NJ method
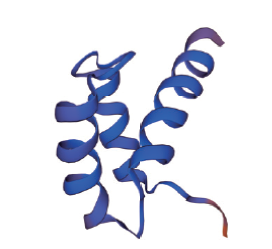
Fig. 2 Prediction diagram of HD tertiary structure of Arabidopsis WOX family proteins This figure is based on the conservative domain of Arabidopsis WOX family, and is predicted using SWISS-MODEL website(https://swissmodel.expasy.org/)
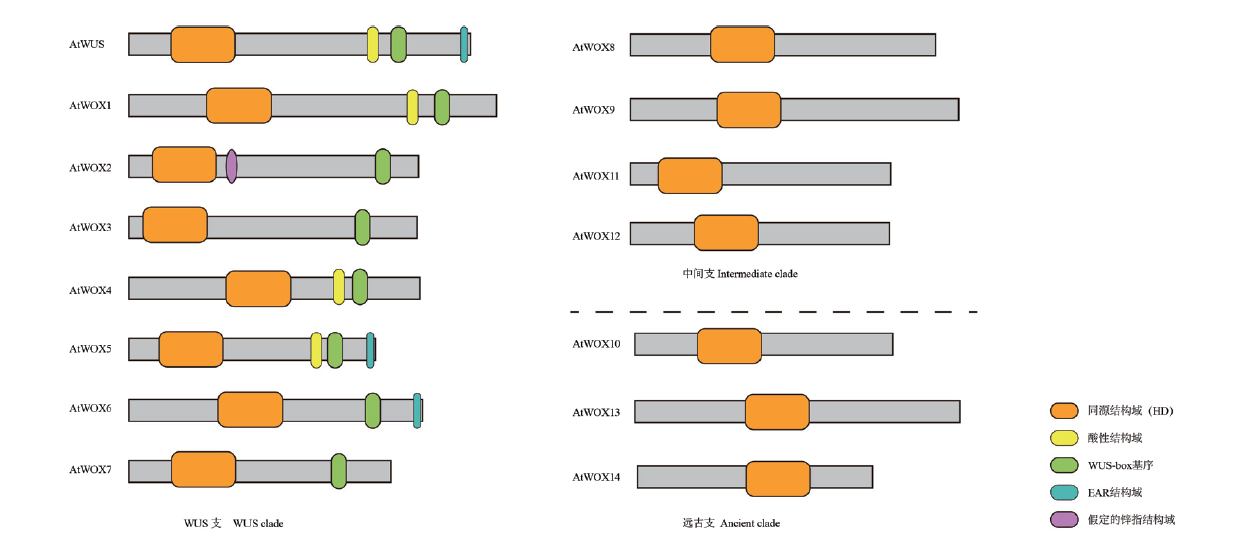
Fig. 3 Schematic diagram of domains of Arabidopsis WOX family proteins Homeodomain(HD)(orange)is the domain shared by members of the family; WUS-box motif(green)is basically in the form of T-L-X-L-F-P-X-X, where X can be any amino acid. There is an assumed zinc finger domain(purple)downstream of HD domain, and only WOX2 has it. The acid domain(yellow)is located at about 10 amino acid residues upstream of WUS box of WUS, WOX1, WOX4 and WOX5. The EAR domain(blue)is strictly defined as L -[ED]- L -[RST]- L. This domain only exists with the carboxyl terminus of WUS, WOX5 and WOX7
| 基因 Genes | 物种 Species | 功能 Function | 参考文献 Reference |
|---|---|---|---|
| WUS | 杨树,小麦 P. tomentosa, and T. aestivum | 初生根和侧根发育,植物株型 Primary root and lateral root development,and plant type | [ |
| WOX1 | 拟南芥,小麦,番茄 A. thaliana, T. aestivum, and L. esculentum | 叶片的侧向生长和形状结构 Lateral growth and shape structure of leaves | [ |
| WOX2 | 拟南芥,挪威云杉,海岸松 A. thaliana, P. abies, and P. pinaster | 胚胎发育 Embryonic development | [ |
| WOX3 | 柳枝稷,拟南芥,水稻,玉米 P. virgatum, A. thaliana, O. sativa, and Z. mays | 叶片发育和形态建成 Leaf development and morphogenesis | [ |
| WOX4 | 拟南芥,水稻 A. thaliana and O. sativa | 促进形成层,初生根的分化 Promote differentiation of cambium and primary root | [ |
| WOX5 | 杨树P. tomentosa | 初生根发育Primary root development | [ |
| WOX6 | 拟南芥A. thaliana | 调控胚珠和种子发育Regulation of ovule and seed development | [ |
| WOX7 | 拟南芥A. thaliana | 侧根发育Lateral root development | [ |
| WOX8 | 拟南芥,水稻 A. thaliana and O. sativa | 胚胎发育,促进主茎生长 Embryonic development, promoting the growth of main stem | [ |
| WOX9 | 拟南芥,烟草,矮牵牛,番茄 A. thaliana, N. tabacum, P. hybrid, and L. esculentum | 胚胎发育,根系发育,SAM,花分生组织发育 Embryonic development, root development, SAM, and floral meristem development | [ |
| WOX10 | 水稻O. sativa | 促进初生根的分化Promote differentiation of primary roots | [ |
| WOX11 | 水稻,人参,杨树 O. sativa, P. ginseng, and P. tomentosa | 促进根,茎,芽发育 Promote the development of roots, stems and buds | [ |
| WOX12 | 拟南芥,杨树A. thaliana and P. tomentosa | 根系发育Root development | [ |
| WOX13 | 拟南芥A. thaliana | 促进器官重生,花器官形成 Promote organ regeneration and flower organ formation | [ |
| WOX14 | 拟南芥A. thaliana | 根和花的发育Development of roots and flowers | [ |
Table 1 WOX transcription factors related to plant growth and development
| 基因 Genes | 物种 Species | 功能 Function | 参考文献 Reference |
|---|---|---|---|
| WUS | 杨树,小麦 P. tomentosa, and T. aestivum | 初生根和侧根发育,植物株型 Primary root and lateral root development,and plant type | [ |
| WOX1 | 拟南芥,小麦,番茄 A. thaliana, T. aestivum, and L. esculentum | 叶片的侧向生长和形状结构 Lateral growth and shape structure of leaves | [ |
| WOX2 | 拟南芥,挪威云杉,海岸松 A. thaliana, P. abies, and P. pinaster | 胚胎发育 Embryonic development | [ |
| WOX3 | 柳枝稷,拟南芥,水稻,玉米 P. virgatum, A. thaliana, O. sativa, and Z. mays | 叶片发育和形态建成 Leaf development and morphogenesis | [ |
| WOX4 | 拟南芥,水稻 A. thaliana and O. sativa | 促进形成层,初生根的分化 Promote differentiation of cambium and primary root | [ |
| WOX5 | 杨树P. tomentosa | 初生根发育Primary root development | [ |
| WOX6 | 拟南芥A. thaliana | 调控胚珠和种子发育Regulation of ovule and seed development | [ |
| WOX7 | 拟南芥A. thaliana | 侧根发育Lateral root development | [ |
| WOX8 | 拟南芥,水稻 A. thaliana and O. sativa | 胚胎发育,促进主茎生长 Embryonic development, promoting the growth of main stem | [ |
| WOX9 | 拟南芥,烟草,矮牵牛,番茄 A. thaliana, N. tabacum, P. hybrid, and L. esculentum | 胚胎发育,根系发育,SAM,花分生组织发育 Embryonic development, root development, SAM, and floral meristem development | [ |
| WOX10 | 水稻O. sativa | 促进初生根的分化Promote differentiation of primary roots | [ |
| WOX11 | 水稻,人参,杨树 O. sativa, P. ginseng, and P. tomentosa | 促进根,茎,芽发育 Promote the development of roots, stems and buds | [ |
| WOX12 | 拟南芥,杨树A. thaliana and P. tomentosa | 根系发育Root development | [ |
| WOX13 | 拟南芥A. thaliana | 促进器官重生,花器官形成 Promote organ regeneration and flower organ formation | [ |
| WOX14 | 拟南芥A. thaliana | 根和花的发育Development of roots and flowers | [ |
| 基因 Genes | 物种 Species | 研究方法 Methods | 胁迫类型 Stress type | 生物学功能 Biological function | 参考文献 Ref. |
|---|---|---|---|---|---|
| OsWOX13 | 水稻 O. sativa | 过表达 Overexpression | 干旱胁迫 Drought | 利用rab21启动子驱动OsWOX13过表达导致水稻耐旱性增强 Overexpression of OsWOX13 driven by rab21 promoter leads to enhanced drought tolerance in rice | [ |
| JcWOX5 | 麻风树基因导入水稻 O. sativa and Jatro-pha curcas | 过表达 Overexpression | 干旱胁迫 Drought | JcWOX5负调控拟南芥的干旱反应 JcWOX5 negatively regulates drought response | [ |
| OsWOX10,QHB/OsWOX5 | 水稻 O. sativa | CRISPR CRISPR-qhb | 干旱胁迫 Drought | QHB和OsWOX10控制土壤条件下轻度干旱诱导LR发育 LR development induced by mild drought in soil controlled by QHB and OsWOX10 | [ |
| GhWOX4 | 棉花,拟南芥中异位表达 G. Hirsutum and A. thaliana | 过表达,病毒诱导基因沉默(VIGS)技术 Overexpression,Virus induced gene silencing technology | 干旱胁迫 Drought | GhWOX4正调控棉花耐旱性 GhWOX4 positively regulated drought tolerance | [ |
| PagWOX11/12a | 杂交白杨84K Pop-ulus alba×Populus glandulosa | 过表达 Overexpression | 干旱胁迫 Drought | PagWOX11/12a通过促进根系伸长,生物量增长,ROS清除相关基因的表达来调节ROS水平,从而提高植物耐旱性 PagWOX11/12a regulates ROS level by promoting root elongation, biomass growth and ROS clearance related gene expression, thus improving plant drought tolerance | [ |
| WOX6 | 拟南芥 A. thaliana | hos9-1突变体 hos9-1 mutant | 冷胁迫 Freezing | HOS9正调控拟南芥对冷胁迫的耐受性 HOS9 positively regulated freezing tolerance | [ |
| WOX11 | 水稻 O. sativa | 过表达 Overexpression | 营养胁迫 Nutritional | WOX11基因能够控制根系发育并提高水稻对缺钾的耐受性 WOX11 can control root development and improve tolerance of rice to potassium deficiency | [ |
| PagWOX11/12a | 杨树 P. tomentosa | 过表达 Overexpression | 盐胁迫 Salt | PagWOX11/12a通过激活PagCYP736A12基因的表达从而正调控杨树的耐盐性 PagWOX11/12a regulates salt tolerance of poplar by activating the expression of PagCYP736A12 gene | [ |
Table 2 WOX transcription factors involved in plant abiotic stress responses
| 基因 Genes | 物种 Species | 研究方法 Methods | 胁迫类型 Stress type | 生物学功能 Biological function | 参考文献 Ref. |
|---|---|---|---|---|---|
| OsWOX13 | 水稻 O. sativa | 过表达 Overexpression | 干旱胁迫 Drought | 利用rab21启动子驱动OsWOX13过表达导致水稻耐旱性增强 Overexpression of OsWOX13 driven by rab21 promoter leads to enhanced drought tolerance in rice | [ |
| JcWOX5 | 麻风树基因导入水稻 O. sativa and Jatro-pha curcas | 过表达 Overexpression | 干旱胁迫 Drought | JcWOX5负调控拟南芥的干旱反应 JcWOX5 negatively regulates drought response | [ |
| OsWOX10,QHB/OsWOX5 | 水稻 O. sativa | CRISPR CRISPR-qhb | 干旱胁迫 Drought | QHB和OsWOX10控制土壤条件下轻度干旱诱导LR发育 LR development induced by mild drought in soil controlled by QHB and OsWOX10 | [ |
| GhWOX4 | 棉花,拟南芥中异位表达 G. Hirsutum and A. thaliana | 过表达,病毒诱导基因沉默(VIGS)技术 Overexpression,Virus induced gene silencing technology | 干旱胁迫 Drought | GhWOX4正调控棉花耐旱性 GhWOX4 positively regulated drought tolerance | [ |
| PagWOX11/12a | 杂交白杨84K Pop-ulus alba×Populus glandulosa | 过表达 Overexpression | 干旱胁迫 Drought | PagWOX11/12a通过促进根系伸长,生物量增长,ROS清除相关基因的表达来调节ROS水平,从而提高植物耐旱性 PagWOX11/12a regulates ROS level by promoting root elongation, biomass growth and ROS clearance related gene expression, thus improving plant drought tolerance | [ |
| WOX6 | 拟南芥 A. thaliana | hos9-1突变体 hos9-1 mutant | 冷胁迫 Freezing | HOS9正调控拟南芥对冷胁迫的耐受性 HOS9 positively regulated freezing tolerance | [ |
| WOX11 | 水稻 O. sativa | 过表达 Overexpression | 营养胁迫 Nutritional | WOX11基因能够控制根系发育并提高水稻对缺钾的耐受性 WOX11 can control root development and improve tolerance of rice to potassium deficiency | [ |
| PagWOX11/12a | 杨树 P. tomentosa | 过表达 Overexpression | 盐胁迫 Salt | PagWOX11/12a通过激活PagCYP736A12基因的表达从而正调控杨树的耐盐性 PagWOX11/12a regulates salt tolerance of poplar by activating the expression of PagCYP736A12 gene | [ |
| [1] |
Laux T, Mayer KF, Berger J, et al. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis[J]. Development, 1996, 122(1): 87-96.
doi: 10.1242/dev.122.1.87 pmid: 8565856 |
| [2] |
Haecker A, Gross-Hardt R, Geiges B, et al. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana[J]. Development, 2004, 131(3): 657-668.
doi: 10.1242/dev.00963 pmid: 14711878 |
| [3] |
Chen SK, Kurdyukov S, Kereszt A, et al. The association of homeobox gene expression with stem cell formation and morphogenesis in cultured Medicago truncatula[J]. Planta, 2009, 230(4): 827-840.
doi: 10.1007/s00425-009-0988-1 pmid: 19639337 |
| [4] |
Zhang X, Zong J, Liu JH, et al. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar[J]. J Integr Plant Biol, 2010, 52(11): 1016-1026.
doi: 10.1111/j.1744-7909.2010.00982.x |
| [5] |
Cheng SF, Huang YL, Zhu N, et al. The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response[J]. Gene, 2014, 549(2): 266-274.
doi: 10.1016/j.gene.2014.08.003 URL |
| [6] |
Li MD, Wang RH, Liu ZY, et al. Genome-wide identification and analysis of the WUSCHEL-related homeobox(WOX)gene family in allotetraploid Brassica napus reveals changes in WOX genes during polyploidization[J]. BMC Genomics, 2019, 20(1): 317.
doi: 10.1186/s12864-019-5684-3 |
| [7] |
Rathour M, Sharma A, Kaur A, et al. Genome-wide characterization and expression and co-expression analysis suggested diverse functions of WOX genes in bread wheat[J]. Heliyon, 2020, 6(12): e05762.
doi: 10.1016/j.heliyon.2020.e05762 URL |
| [8] |
Shafique Khan F, Zeng RF, Gan ZM, et al. Genome-wide identification and expression profiling of the WOX gene family in Citrus sinensis and functional analysis of a CsWUS member[J]. Int J Mol Sci, 2021, 22(9): 4919.
doi: 10.3390/ijms22094919 URL |
| [9] |
van der Graaff E, Laux T, Rensing SA. The WUS homeobox-containing(WOX)protein family[J]. Genome Biol, 2009, 10(12): 248.
doi: 10.1186/gb-2009-10-12-248 pmid: 20067590 |
| [10] |
Nardmann J, Reisewitz P, Werr W. Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms[J]. Mol Biol Evol, 2009, 26(8): 1745-1755.
doi: 10.1093/molbev/msp084 pmid: 19387013 |
| [11] |
Mukherjee K, Brocchieri L, Bürglin TR. A comprehensive classification and evolutionary analysis of plant homeobox genes[J]. Mol Biol Evol, 2009, 26(12): 2775-2794.
doi: 10.1093/molbev/msp201 pmid: 19734295 |
| [12] |
Alvarez JM, Bueno N, Cañas RA, et al. Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in Pinus pinaster: new insights into the gene family evolution[J]. Plant Physiol Biochem, 2018, 123: 304-318.
doi: 10.1016/j.plaphy.2017.12.031 URL |
| [13] |
Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning[J]. Plant Cell, 2009, 21(11): 3493-3505.
doi: 10.1105/tpc.109.069997 URL |
| [14] |
Paponov IA, Teale W, Lang D, et al. The evolution of nuclear auxin signalling[J]. BMC Evol Biol, 2009, 9: 126.
doi: 10.1186/1471-2148-9-126 pmid: 19493348 |
| [15] |
Deveaux Y, Toffano-Nioche C, Claisse G, et al. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis[J]. BMC Evol Biol, 2008, 8: 291.
doi: 10.1186/1471-2148-8-291 pmid: 18950478 |
| [16] |
Kong DY, Hao YL, Cui HC. The WUSCHEL related homeobox protein WOX7 regulates the sugar response of lateral root development in Arabidopsis thaliana[J]. Mol Plant, 2016, 9(2): 261-270.
doi: 10.1016/j.molp.2015.11.006 URL |
| [17] |
Xiao W, Molina D, Wunderling A, et al. Pluripotent pericycle cells trigger different growth outputs by integrating developmental cues into distinct regulatory modules[J]. Curr Biol, 2020, 30(22): 4384-4398.e5.
doi: 10.1016/j.cub.2020.08.053 URL |
| [18] |
Li Z, Liu D, Xia Y, et al. Identification of the WUSCHEL-related homeobox(WOX)gene family, and interaction and functional analysis of TaWOX9 and TaWUS in wheat[J]. Int J Mol Sci, 2020, 21(5): 1581.
doi: 10.3390/ijms21051581 URL |
| [19] |
Zhou SL, Jiang W, Long F, et al. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem[J]. Plant Cell, 2017, 29(5): 1088-1104.
doi: 10.1105/tpc.16.00908 URL |
| [20] |
Chen RR, Xu N, Yu B, et al. The WUSCHEL-related homeobox transcription factor OsWOX4 controls the primary root elongation by activating OsAUX1 in rice[J]. Plant Sci, 2020, 298: 110575.
doi: 10.1016/j.plantsci.2020.110575 URL |
| [21] |
Kawai T, Shibata K, Akahoshi R, et al. WUSCHEL-related homeobox family genes in rice control lateral root primordium size[J]. Proc Natl Acad Sci USA, 2022, 119(1): e2101846119.
doi: 10.1073/pnas.2101846119 URL |
| [22] |
Li JB, Zhang J, Jia HX, et al. The WUSCHEL-related homeobox 5a(PtoWOX5a)is involved in adventitious root development in poplar[J]. Tree Physiol, 2018, 38(1): 139-153.
doi: 10.1093/treephys/tpx118 URL |
| [23] |
Li JB, Jia HX, Sun P, et al. The WUSCHELa(PtoWUSa)is involved in developmental plasticity of adventitious root in poplar[J]. Genes, 2020, 11(2): 176.
doi: 10.3390/genes11020176 URL |
| [24] |
Liu J, Chen T, Zhang J, et al. Ginsenosides regulate adventitious root formation in Panax ginseng via a CLE45-WOX11 regulatory module[J]. J Exp Bot, 2020, 71(20): 6396-6407.
doi: 10.1093/jxb/eraa375 URL |
| [25] |
Wu XL, Dabi T, Weigel D. Requirement of homeobox gene STIMPY-/WOX9 for Arabidopsis meristem growth and maintenance[J]. Curr Biol, 2005, 15(5): 436-440.
doi: 10.1016/j.cub.2004.12.079 URL |
| [26] |
Mauriat M, Moritz T. Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation[J]. Plant J, 2009, 58(6): 989-1003.
doi: 10.1111/tpj.2009.58.issue-6 URL |
| [27] |
Israelsson M, Sundberg B, Moritz T. Tissue-specific localization of gibberellins and expression of gibberellin-biosynthetic and signaling genes in wood-forming tissues in aspen[J]. Plant J, 2005, 44(3): 494-504.
pmid: 16236158 |
| [28] |
Denis E, Kbiri N, Mary V, et al. WOX14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis[J]. Plant J, 2017, 90(3): 560-572.
doi: 10.1111/tpj.13513 URL |
| [29] |
Dai XH, Wang J, Song YG, et al. Cytosine methylation of the FWA promoter promotes direct in vitro shoot regeneration in Arabidopsis thaliana[J]. J Integr Plant Biol, 2021, 63(8): 1491-1504.
doi: 10.1111/jipb.v63.8 URL |
| [30] |
Ikeuchi M, Iwase A, Ito T, et al. Wound-inducible WUSCHEL-RELATED HOMEOBOX 13 is required for callus growth and organ reconnection[J]. Plant Physiol, 2022, 188(1): 425-441.
doi: 10.1093/plphys/kiab510 URL |
| [31] |
Wang WF, Li G, Zhao J, et al. Dwarf Tiller1, a Wuschel-related homeobox transcription factor, is required for tiller growth in rice[J]. PLoS Genet, 2014, 10(3): e1004154.
doi: 10.1371/journal.pgen.1004154 URL |
| [32] |
Cheng SF, Tan F, Lu Y, et al. WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice[J]. Nucleic Acids Res, 2018, 46(5): 2356-2369.
doi: 10.1093/nar/gky017 URL |
| [33] |
Si XM, Wang WX, Wang K, et al. A sheathed spike gene, TaWUS-like inhibits stem elongation in common wheat by regulating hormone levels[J]. Int J Mol Sci, 2021, 22(20): 11210.
doi: 10.3390/ijms222011210 URL |
| [34] |
Wang H, Niu LF, Fu CX, et al. Overexpression of the WOX gene STENOFOLIA improves biomass yield and sugar release in transgenic grasses and display altered cytokinin homeostasis[J]. PLoS Genet, 2017, 13(3): e1006649.
doi: 10.1371/journal.pgen.1006649 URL |
| [35] |
Yang RJ, Wu ZY, Bai C, et al. Overexpression of PvWOX3a in switchgrass promotes stem development and increases plant height[J]. Hortic Res, 2021, 8(1): 252.
doi: 10.1038/s41438-021-00678-w |
| [36] |
Liu BB, Zhang J, Yang ZH, et al. PtWOX11 acts as master regulator conducting the expression of key transcription factors to induce de novo shoot organogenesis in poplar[J]. Plant Mol Biol, 2018, 98(4/5): 389-406.
doi: 10.1007/s11103-018-0786-x |
| [37] |
Zhang ZJ, Runions A, Mentink RA, et al. A WOX/auxin biosynthesis module controls growth to shape leaf form[J]. Curr Biol, 2020, 30(24): 4857-4868.e6.
doi: 10.1016/j.cub.2020.09.037 URL |
| [38] |
Dai MQ, Hu YF, Zhao Y, et al. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development[J]. Plant Physiol, 2007, 144(1): 380-390.
doi: 10.1104/pp.107.095737 URL |
| [39] |
Honda E, Yew CL, Yoshikawa T, et al. Leaf lateral symmetry1, a member of the wuschel-related homeobox3 gene family, regulates lateral organ development differentially from other paralogs, narrow leaf2 and narrow leaf3 in rice[J]. Plant Cell Physiol, 2018, 59(2): 376-391.
doi: 10.1093/pcp/pcx196 URL |
| [40] |
Uzair M, Long HX, Zafar SA, et al. Narrow Leaf21, encoding ribosomal protein RPS3A, controls leaf development in rice[J]. Plant Physiol, 2021, 186(1): 497-518.
doi: 10.1093/plphys/kiab075 pmid: 33591317 |
| [41] |
Yasui Y, Ohmori Y, Takebayashi Y, et al. WUSCHEL-RELATED HOMEOBOX4 acts as a key regulator in early leaf development in rice[J]. PLoS Genet, 2018, 14(4): e1007365.
doi: 10.1371/journal.pgen.1007365 URL |
| [42] |
Liu MY, Lei L, Miao F, et al. The STENOFOLIA gene from Medicago alters leaf width, flowering time and chlorophyll content in transgenic wheat[J]. Plant Biotechnol J, 2018, 16(1): 186-196.
doi: 10.1111/pbi.2018.16.issue-1 URL |
| [43] |
Wolabu TW, Wang H, Tadesse D, et al. WOX9 functions antagonistic to STF and LAM1 to regulate leaf blade expansion in Medicago truncatula and Nicotiana sylvestris[J]. New Phytol, 2021, 229(3): 1582-1597.
doi: 10.1111/nph.v229.3 URL |
| [44] |
Nardmann J, Ji JB, Werr W, et al. The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems[J]. Development, 2004, 131(12): 2827-2839.
doi: 10.1242/dev.01164 pmid: 15169755 |
| [45] |
Yoshikawa T, Tanaka SY, Masumoto Y, et al. Barley narrow leafed dwarf1 encoding a wuschel-related homeobox 3(wox3)regulates the marginal development of lateral organs[J]. Breed Sci, 2016, 66(3): 416-424.
doi: 10.1270/jsbbs.16019 URL |
| [46] | Conklin PA, Johnston R, Conlon BR, et al. Plant homeodomain proteins provide a mechanism for how leaves grow wide[J]. Development, 2020, 147(20): dev193623. |
| [47] |
Kong DX, Pan X, Jing YF, et al. ZmSPL10/14/26 are required for epidermal hair cell fate specification on maize leaf[J]. New Phytol, 2021, 230(4): 1533-1549.
doi: 10.1111/nph.17293 pmid: 33626179 |
| [48] |
Zhang CL, Wang JF, Wang X, et al. UF, a WOX gene, regulates a novel phenotype of un-fused flower in tomato[J]. Plant Sci, 2020, 297: 110523.
doi: 10.1016/j.plantsci.2020.110523 URL |
| [49] |
Du F, Mo YJ, Israeli A, et al. Leaflet initiation and blade expansion are separable in compound leaf development[J]. Plant J, 2020, 104(4): 1073-1087.
doi: 10.1111/tpj.v104.4 URL |
| [50] |
Wang CQ, Zhao BL, He LL, et al. The WOX family transcriptional regulator SlLAM1 controls compound leaf and floral organ development in Solanum lycopersicum[J]. J Exp Bot, 2021, 72(5): 1822-1835.
doi: 10.1093/jxb/eraa574 URL |
| [51] |
Nakayama H, Rowland SD, Cheng ZZ, et al. Leaf form diversification in an ornamental heirloom tomato results from alterations in two different HOMEOBOX genes[J]. Curr Biol, 2021, 31(21): 4788-4799.e5.
doi: 10.1016/j.cub.2021.08.023 URL |
| [52] |
Matsumoto N, Okada K. A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers[J]. Genes Dev, 2001, 15(24): 3355-3364.
doi: 10.1101/gad.931001 URL |
| [53] | Minh-Thu PT, Kim JS, Chae S, et al. A WUSCHEL homeobox transcription factor, OsWOX13, enhances drought tolerance and triggers early flowering in rice[J]. Mol Cells, 2018, 41(8): 781-798. |
| [54] |
Rebocho AB, Bliek M, Kusters E, et al. Role of EVERGREEN in the development of the cymose Petunia inflorescence[J]. Dev Cell, 2008, 15(3): 437-447.
doi: 10.1016/j.devcel.2008.08.007 URL |
| [55] |
Park SO, Hwang S, Hauser BA. The phenotype of Arabidopsis ovule mutants mimics the morphology of primitive seed plants[J]. Proc Biol Sci, 2004, 271(1536): 311-316.
doi: 10.1098/rspb.2003.2544 URL |
| [56] |
Park SO, Zheng ZG, Oppenheimer DG, et al. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development[J]. Development, 2005, 132(4): 841-849.
doi: 10.1242/dev.01654 URL |
| [57] |
Gu R, Song XF, Liu XF, et al. Genome-wide analysis of CsWOX transcription factor gene family in cucumber(Cucumis sativus L.)[J]. Sci Rep, 2020, 10(1): 6216.
doi: 10.1038/s41598-020-63197-z |
| [58] |
Wu XL, Chory J, Weigel D. Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development[J]. Dev Biol, 2007, 309(2): 306-316.
doi: 10.1016/j.ydbio.2007.07.019 URL |
| [59] |
Breuninger H, Rikirsch E, Hermann M, et al. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo[J]. Dev Cell, 2008, 14(6): 867-876.
doi: 10.1016/j.devcel.2008.03.008 pmid: 18539115 |
| [60] |
Samakovli D, Tichá T, Vavrdová T, et al. HEAT SHOCK PROTEIN 90 proteins and YODA regulate main body axis formation during early embryogenesis[J]. Plant Physiol, 2021, 186(3): 1526-1544.
doi: 10.1093/plphys/kiab171 pmid: 33856486 |
| [61] |
Zhu TQ, Moschou PN, Alvarez JM, et al. WUSCHEL-RELATED HOMEOBOX 2 is important for protoderm and suspensor development in the gymnosperm Norway spruce[J]. BMC Plant Biol, 2016, 16: 19.
doi: 10.1186/s12870-016-0706-7 URL |
| [62] |
Hassani SB, Trontin JF, Raschke J, et al. Constitutive overexpression of a conifer WOX2 homolog affects somatic embryo development in Pinus pinaster and promotes somatic embryogenesis and organogenesis in Arabidopsis seedlings[J]. Front Plant Sci, 2022, 13: 838421.
doi: 10.3389/fpls.2022.838421 URL |
| [63] |
Hendelman A, Zebell S, Rodriguez-Leal D, et al. Conserved pleiotropy of an ancient plant homeobox gene uncovered by Cis-regulatory dissection[J]. Cell, 2021, 184(7): 1724-1739.e16.
doi: 10.1016/j.cell.2021.02.001 pmid: 33667348 |
| [64] |
Tang YH, Li H, Guan YX, et al. Genome-wide identification of the physic nut WUSCHEL-related homeobox gene family and functional analysis of the abiotic stress responsive gene JcWOX5[J]. Front Genet, 2020, 11: 670.
doi: 10.3389/fgene.2020.00670 URL |
| [65] |
Sajjad M, Wei X, Liu LS, et al. Transcriptome analysis revealed GhWOX4 intercedes myriad regulatory pathways to modulate drought tolerance and vascular growth in cotton[J]. Int J Mol Sci, 2021, 22(2): 898.
doi: 10.3390/ijms22020898 URL |
| [66] | Wang LQ, Li Z, Wen SS, et al. WUSCHEL-related homeobox gene PagWOX11/12a responds to drought stress by enhancing root elongation and biomass growth in poplar[J]. J Exp Bot, 2020, 71(4): 1503-1513. |
| [67] |
Liu R, Wang R, Lu MZ, et al. WUSCHEL-related homeobox gene PagWOX11/12a is involved in drought tolerance through modulating reactive oxygen species scavenging in poplar[J]. Plant Signal Behav, 2021, 16(3): 1866312.
doi: 10.1080/15592324.2020.1866312 URL |
| [68] |
Zhu JH, Shi HZ, Lee BH, et al. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway[J]. Proc Natl Acad Sci USA, 2004, 101(26): 9873-9878.
doi: 10.1073/pnas.0403166101 URL |
| [69] |
Chen G, Feng HM, Hu QD, et al. Improving rice tolerance to potassium deficiency by enhancing OsHAK16p: WOX11-controlled root development[J]. Plant Biotechnol J, 2015, 13(6): 833-848.
doi: 10.1111/pbi.2015.13.issue-6 URL |
| [70] |
Wang LQ, Wen SS, Wang R, et al. PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar[J]. Plant Biotechnol J, 2021, 19(11): 2249-2260.
doi: 10.1111/pbi.v19.11 URL |
| [1] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [2] | HU Hai-lin, XU Li, LI Xiao-xu, WANG Chen-can, MEI Man, DING Wen-jing, ZHAO Yuan-yuan. Advances in the Regulation of Plant Growth, Development and Stress Physiology by Small Peptide Hormones [J]. Biotechnology Bulletin, 2023, 39(7): 13-25. |
| [3] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [4] | LI Yuan-hong, GUO Yu-hao, CAO Yan, ZHU Zhen-zhou, WANG Fei-fei. Research Progress in the Microalgal Growth and Accumulation of Target Products Regulated by Exogenous Phytohormone [J]. Biotechnology Bulletin, 2023, 39(6): 61-72. |
| [5] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| [6] | XUE Jiao ZHU Qing-feng FENG Yan-zhao CHEN Pei LIU Wen-hua ZHANG Ai-xia LIU Qin-jian ZHANG Qi YU Yang. Advances in Upstream Open Reading Frame in Plant Genes [J]. Biotechnology Bulletin, 2023, 39(4): 157-165. |
| [7] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| [8] | WEI Ming WANG Xin-yu WU Guo-qiang ZHAO Meng. The Role of NAD-dependent Deacetylase SRT in Plant Epigenetic Inheritance Regulation [J]. Biotechnology Bulletin, 2023, 39(4): 59-70. |
| [9] | SANG Tian, WANG Peng-cheng. Research Progress in Plant SUMOylation [J]. Biotechnology Bulletin, 2023, 39(3): 1-12. |
| [10] | YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development [J]. Biotechnology Bulletin, 2023, 39(3): 133-142. |
| [11] | ZHAO Meng-liang, GUO Yi-ting, REN Yan-jing. Identification and Analysis of WRKY Transcription Factor Family Genes in Helianthus tuberosus [J]. Biotechnology Bulletin, 2023, 39(2): 116-125. |
| [12] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [13] | XU Rui, ZHU Ying-fang. The Key Roles of Mediator Complex in Plant Responses to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 54-60. |
| [14] | CHEN Guang-xia, LI Xiu-jie, JIANG Xi-long, SHAN Lei, ZHANG Zhi-chang, LI Bo. Research Progress in Plant Small Signaling Peptides Involved in Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(11): 61-73. |
| [15] | HAN Fang-ying, HU Xin, WANG Nan-nan, XIE Yu-hong, WANG Xiao-yan, ZHU Qiang. Research Progress in Response of DREBs to Abiotic Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(11): 86-98. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||