








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (6): 233-247.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1369
Previous Articles Next Articles
YIN Ming-hua1,2,3,4( ), YU Huan-yuan1, XIAO Xin-yi1, WANG Yu-ting1
), YU Huan-yuan1, XIAO Xin-yi1, WANG Yu-ting1
Received:2022-11-07
Online:2023-06-26
Published:2023-07-07
Contact:
YIN Ming-hua
E-mail:yinminghua04@163.com
YIN Ming-hua, YU Huan-yuan, XIAO Xin-yi, WANG Yu-ting. Chloroplast Genomic Characterization and Phylogenetic Analysis of Colocasia esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan[J]. Biotechnology Bulletin, 2023, 39(6): 233-247.
| 基因分类 Category | 基因分组 Gene group | 基因名称 Gene name |
|---|---|---|
| 光合作用 Photosynthesis | 光合系统I基因 Photosystem I | psaA、psaB、psaC、psaI、psaJ |
| 光合系统II基因 Photosystem II | psbA、psbB、psbC、psbD、psbE、psbF、psbH、psbI、psbK、psbL、psbM、psbT、psbZ | |
| 细胞色素复合物基因 Cytochrome b/f complex | petA、petB、petD、petG、petL、petN | |
| ATP合成酶基因 ATP synthase | atpA、atpB、atpE、atpF、atpH、atpI | |
| NADH脱氢酶基因 NADH dehydrogenase | ndhA、ndhB(2)、ndhC、ndhD、ndhE、ndhF、ndhG、ndhH、ndhI、ndhJ、ndhK | |
| 二磷酸核酮糖羧化酶大亚基基因 Rubisco large subunit | rbcL | |
| 自我复制Self-replication | RNA聚合酶亚基基因RNA polymeras | rpoA、rpoB、rpoC1、rpoC2 |
| 核糖体小亚基基因 Ribosomal proteins(SSU) | rps11、rps12(2)、rps14、rps15、rps16、rps18、 rps19、rps2、rps3、rps4、rps7(2)、rps8 | |
| 核糖体大亚基基因 Ribosomal proteins(LSU) | rpl14、rpl16、rpl2(2)、rpl20、rpl22、rpl23(2)、rpl32、rpl33、rpl36、 | |
| 转运RNA基因 Transfer RNAs | trnC-GCA、trnD-GUC、trnF-GAA、trnG-GCC、trnH-GUG、trnI-CAU(2)、trnK-UUU、trnL-CAA(2)、trnL-UAA、trnL-UAG、trnM-CAU、trnN-GUU(2)、trnP-UGG、trnQ-UUG、trnR-ACG(2)、trnR-UCU、trnS-GCU、trnS-GGA、trnS-UGA、trnT-UGU、trnV-GAC(2)、trnV-UAC、trnW-CCA、trnY-GUA、trnfM-CAU | |
| 核糖体RNA基因 Ribosomal RNAs | rrn16(2)、rrn23(2)、rrn4.5(2)、rrn5(2) | |
| 其他基因 Other genes | 成熟酶基因 Maturase | matK |
| 蛋白酶Protease | clpP1 | |
| 乙酰辅酶 A 羧化酶Acetyl-CoA carboxylase | accD | |
| 翻译起始因子 Translation initiation factor | infA | |
| 包膜蛋白基因 Envelope membrane protein | cemA | |
| C型细胞色素合成基因 C-type cytochrome synthesis gene | ccsA | |
| 未知功能Unknown function | 假定叶绿体阅读框 Hypothetical chloroplast reading frames | ycf1、ycf2(2) |
Table 1 List of genes found in chloroplast genome of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
| 基因分类 Category | 基因分组 Gene group | 基因名称 Gene name |
|---|---|---|
| 光合作用 Photosynthesis | 光合系统I基因 Photosystem I | psaA、psaB、psaC、psaI、psaJ |
| 光合系统II基因 Photosystem II | psbA、psbB、psbC、psbD、psbE、psbF、psbH、psbI、psbK、psbL、psbM、psbT、psbZ | |
| 细胞色素复合物基因 Cytochrome b/f complex | petA、petB、petD、petG、petL、petN | |
| ATP合成酶基因 ATP synthase | atpA、atpB、atpE、atpF、atpH、atpI | |
| NADH脱氢酶基因 NADH dehydrogenase | ndhA、ndhB(2)、ndhC、ndhD、ndhE、ndhF、ndhG、ndhH、ndhI、ndhJ、ndhK | |
| 二磷酸核酮糖羧化酶大亚基基因 Rubisco large subunit | rbcL | |
| 自我复制Self-replication | RNA聚合酶亚基基因RNA polymeras | rpoA、rpoB、rpoC1、rpoC2 |
| 核糖体小亚基基因 Ribosomal proteins(SSU) | rps11、rps12(2)、rps14、rps15、rps16、rps18、 rps19、rps2、rps3、rps4、rps7(2)、rps8 | |
| 核糖体大亚基基因 Ribosomal proteins(LSU) | rpl14、rpl16、rpl2(2)、rpl20、rpl22、rpl23(2)、rpl32、rpl33、rpl36、 | |
| 转运RNA基因 Transfer RNAs | trnC-GCA、trnD-GUC、trnF-GAA、trnG-GCC、trnH-GUG、trnI-CAU(2)、trnK-UUU、trnL-CAA(2)、trnL-UAA、trnL-UAG、trnM-CAU、trnN-GUU(2)、trnP-UGG、trnQ-UUG、trnR-ACG(2)、trnR-UCU、trnS-GCU、trnS-GGA、trnS-UGA、trnT-UGU、trnV-GAC(2)、trnV-UAC、trnW-CCA、trnY-GUA、trnfM-CAU | |
| 核糖体RNA基因 Ribosomal RNAs | rrn16(2)、rrn23(2)、rrn4.5(2)、rrn5(2) | |
| 其他基因 Other genes | 成熟酶基因 Maturase | matK |
| 蛋白酶Protease | clpP1 | |
| 乙酰辅酶 A 羧化酶Acetyl-CoA carboxylase | accD | |
| 翻译起始因子 Translation initiation factor | infA | |
| 包膜蛋白基因 Envelope membrane protein | cemA | |
| C型细胞色素合成基因 C-type cytochrome synthesis gene | ccsA | |
| 未知功能Unknown function | 假定叶绿体阅读框 Hypothetical chloroplast reading frames | ycf1、ycf2(2) |
| 区域Area | 碱基长度 Base length/ bp | T/% | C/% | A/% | G/% | GC/% | AT偏斜值AT skew | GC 偏斜值GC skew |
|---|---|---|---|---|---|---|---|---|
| 大单拷贝区LSC | 89 814 | 33.58 | 17.59 | 32.04 | 16.79 | 34.38 | -0.023 5 | -0.023 3 |
| 小单拷贝区SSC | 22 074 | 36.02 | 15.12 | 34.97 | 13.89 | 29.01 | -0.014 8 | -0.042 4 |
| 反向重复序列a(IRa) | 25 328 | 29.01 | 21.95 | 28.59 | 20.44 | 42.40 | -0.007 3 | -0.035 6 |
| 反向重复序列b(IRb) | 25 328 | 28.59 | 20.44 | 29.01 | 21.95 | 42.40 | 0.007 3 | 0.035 6 |
| 总基因组 | 162 544 | 32.42 | 18.38 | 31.43 | 17.77 | 36.15 | -0.015 5 | -0.016 9 |
Table 2 Nucleotide composition of the whole chloroplast genome of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
| 区域Area | 碱基长度 Base length/ bp | T/% | C/% | A/% | G/% | GC/% | AT偏斜值AT skew | GC 偏斜值GC skew |
|---|---|---|---|---|---|---|---|---|
| 大单拷贝区LSC | 89 814 | 33.58 | 17.59 | 32.04 | 16.79 | 34.38 | -0.023 5 | -0.023 3 |
| 小单拷贝区SSC | 22 074 | 36.02 | 15.12 | 34.97 | 13.89 | 29.01 | -0.014 8 | -0.042 4 |
| 反向重复序列a(IRa) | 25 328 | 29.01 | 21.95 | 28.59 | 20.44 | 42.40 | -0.007 3 | -0.035 6 |
| 反向重复序列b(IRb) | 25 328 | 28.59 | 20.44 | 29.01 | 21.95 | 42.40 | 0.007 3 | 0.035 6 |
| 总基因组 | 162 544 | 32.42 | 18.38 | 31.43 | 17.77 | 36.15 | -0.015 5 | -0.016 9 |
| 核苷酸类型 Nucleotide type | 简单重复序列 Simple sequence repeat | 重复序列个数 Number of SSR |
|---|---|---|
| 单核苷酸 Mononucleotide | A | 45 |
| C | 2 | |
| G | 1 | |
| T | 36 | |
| 二核苷酸 Dinucleotide | AT | 5 |
| TA | 10 | |
| 三核苷酸 Trinucleotide | TTA | 1 |
| 四核苷酸 Tetranucleotide | TATG | 1 |
Table 3 Simple sequence repeats(SSR)type of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
| 核苷酸类型 Nucleotide type | 简单重复序列 Simple sequence repeat | 重复序列个数 Number of SSR |
|---|---|---|
| 单核苷酸 Mononucleotide | A | 45 |
| C | 2 | |
| G | 1 | |
| T | 36 | |
| 二核苷酸 Dinucleotide | AT | 5 |
| TA | 10 | |
| 三核苷酸 Trinucleotide | TTA | 1 |
| 四核苷酸 Tetranucleotide | TATG | 1 |

Fig. 2 IR boundary analysis of the chloroplast genomes of C. cormosus cv. L. Schoot var. Cormosus ‘Hongyayu’ from Jiangxi Yanshan and its 11 related species

Fig. 4 Nucleotide diversity of chloroplast genomes of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan and its 11 related species
| 基因 Gene | GC含量 GC content/% | 基因 Gene | GC含量 GC content/% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 平均 GCall | 第一密码子GC1 | 第二密码子GC2 | 第三密码子GC3 | 有效密码子数ENC | 平均 GCall | 第一密码子GC1 | 第二密码子GC2 | 第三密码子GC3 | 有效密码子数ENC | ||||
| accD | 34.62 | 38.62 | 37.40 | 27.85 | 44.206 | psbH | 42.34 | 48.65 | 47.30 | 31.08 | 43.164 | ||
| atpA | 40.03 | 55.71 | 39.17 | 25.20 | 44.586 | psbI | 36.94 | 48.65 | 29.73 | 32.43 | 40.922 | ||
| atpB | 42.48 | 56.91 | 41.68 | 28.86 | 48.495 | psbJ | 41.46 | 41.46 | 53.66 | 29.27 | 20.769 | ||
| atpE | 41.67 | 52.94 | 41.91 | 30.15 | 46.888 | psbK | 37.57 | 42.86 | 36.51 | 33.33 | 42.160 | ||
| atpF | 37.73 | 50.00 | 35.16 | 28.02 | 39.808 | psbL | 29.06 | 35.90 | 28.21 | 23.08 | 50.514 | ||
| atpH | 44.31 | 62.20 | 48.78 | 21.95 | 49.183 | psbM | 28.57 | 42.86 | 25.71 | 17.14 | 31.500 | ||
| atpI | 38.04 | 49.19 | 37.90 | 27.02 | 43.152 | psbT | 35.29 | 41.18 | 35.29 | 29.41 | 34.772 | ||
| ccsA | 32.1 | 32.10 | 37.96 | 26.23 | 43.768 | psbZ | 34.39 | 38.10 | 42.86 | 22.22 | 51.110 | ||
| cemA | 31.45 | 39.62 | 27.04 | 27.67 | 40.639 | rbcL | 43.04 | 57.38 | 43.66 | 28.07 | 47.314 | ||
| clpP1 | 42.69 | 59.61 | 36.95 | 31.53 | 59.229 | rpl2 | 43.80 | 50.73 | 49.27 | 31.39 | 50.648 | ||
| matK | 31.58 | 38.99 | 31.19 | 24.56 | 46.593 | rpl2 | 43.80 | 50.73 | 49.27 | 31.39 | 50.648 | ||
| ndhA | 35.81 | 44.51 | 37.91 | 25.00 | 44.550 | rpl14 | 39.02 | 53.66 | 38.21 | 25.20 | 56.951 | ||
| ndhB | 37.77 | 42.07 | 39.53 | 31.70 | 47.366 | rpl16 | 43.87 | 50.74 | 53.68 | 27.21 | 40.180 | ||
| ndhB | 37.77 | 42.07 | 39.53 | 31.70 | 47.366 | rpl20 | 36.44 | 37.29 | 44.92 | 27.12 | 43.650 | ||
| ndhC | 37.47 | 49.59 | 33.88 | 28.93 | 45.634 | rpl22 | 37.63 | 45.16 | 40.32 | 27.42 | 51.065 | ||
| ndhD | 35.46 | 39.72 | 38.52 | 28.14 | 49.596 | rpl23 | 37.04 | 41.11 | 40.00 | 30.00 | 52.839 | ||
| ndhE | 31.37 | 38.24 | 33.33 | 22.55 | 42.469 | rpl23 | 37.04 | 41.11 | 40.00 | 30.00 | 52.839 | ||
| ndhF | 33.06 | 37.42 | 36.88 | 24.90 | 45.696 | rpl32 | 33.91 | 34.48 | 44.83 | 22.41 | 34.549 | ||
| ndhG | 33.15 | 42.37 | 33.33 | 23.73 | 43.846 | rpl33 | 34.33 | 38.81 | 38.81 | 25.37 | 42.754 | ||
| ndhH | 38.16 | 51.78 | 35.79 | 26.90 | 48.229 | rpl36 | 37.72 | 42.11 | 47.37 | 23.68 | 46.750 | ||
| ndhI | 34.44 | 43.89 | 36.67 | 22.78 | 44.213 | rpoA | 35.00 | 45.59 | 34.41 | 25.00 | 49.552 | ||
| ndhJ | 39.2 | 48.43 | 37.74 | 31.45 | 51.139 | rpoB | 39.05 | 50.23 | 37.74 | 29.17 | 49.169 | ||
| ndhK | 38.82 | 43.37 | 43.37 | 29.72 | 51.528 | rpoC1 | 38.40 | 50.29 | 38.16 | 26.75 | 49.009 | ||
| pafI | 39.45 | 47.34 | 40.83 | 30.18 | 55.939 | rpoC2 | 37.48 | 46.02 | 37.63 | 28.80 | 50.022 | ||
| pafII | 38.92 | 44.86 | 40.54 | 31.35 | 51.331 | rps2 | 38.96 | 43.88 | 43.04 | 29.96 | 45.542 | ||
| pbf1 | 46.21 | 52.27 | 43.18 | 43.18 | 48.475 | rps3 | 34.86 | 46.58 | 34.25 | 23.74 | 45.101 | ||
| petA | 40.5 | 53.58 | 35.51 | 32.4 | 51.521 | rps4 | 37.79 | 49.01 | 38.12 | 26.24 | 50.288 | ||
| petB | 39.66 | 47.22 | 41.67 | 30.09 | 42.013 | rps7 | 39.10 | 52.56 | 44.87 | 19.87 | 44.746 | ||
| petD | 38.72 | 50.93 | 39.13 | 26.09 | 44.396 | rps7 | 39.10 | 52.56 | 44.87 | 19.87 | 44.746 | ||
| petG | 38.6 | 55.26 | 31.58 | 28.95 | 33.100 | rps8 | 36.09 | 41.35 | 42.11 | 24.81 | 42.540 | ||
| petL | 37.5 | 37.50 | 46.88 | 28.12 | 35.182 | rps11 | 44.12 | 55.40 | 57.55 | 19.42 | 41.507 | ||
| petN | 41.11 | 50.00 | 43.33 | 30.00 | 30.698 | rps12 | 41.94 | 52.42 | 48.39 | 25.00 | 43.732 | ||
| psaA | 43.01 | 51.93 | 43.68 | 33.42 | 49.161 | rps12 | 41.94 | 52.42 | 48.39 | 25.00 | 43.732 | ||
| psaB | 40.91 | 48.44 | 42.99 | 31.29 | 48.022 | rps14 | 41.58 | 42.57 | 46.53 | 35.64 | 37.716 | ||
| psaC | 43.9 | 43.90 | 53.66 | 34.15 | 48.358 | rps15 | 34.05 | 40.86 | 29.03 | 32.26 | 49.599 | ||
| psaI | 35.14 | 43.24 | 27.03 | 35.14 | 44.365 | rps16 | 35.86 | 50.63 | 36.71 | 20.25 | 42.450 | ||
| psaJ | 35.66 | 41.86 | 39.53 | 25.58 | 42.629 | rps18 | 34.31 | 35.29 | 44.12 | 23.53 | 37.936 | ||
| psbA | 42.66 | 49.72 | 43.79 | 34.46 | 41.107 | rps19 | 35.84 | 43.01 | 39.78 | 24.73 | 42.905 | ||
| psbB | 43.42 | 54.42 | 46.37 | 29.47 | 48.296 | ycf1 | 30.85 | 37.27 | 30.03 | 25.25 | 47.451 | ||
| psbC | 44.37 | 54.33 | 46.54 | 32.25 | 45.499 | ycf2 | 37.43 | 41.19 | 34.60 | 36.51 | 52.960 | ||
| psbD | 42.18 | 51.98 | 43.50 | 31.07 | 44.142 | ycf2 | 37.43 | 41.19 | 34.60 | 36.51 | 52.960 | ||
| psbE | 40.08 | 42.86 | 46.43 | 30.95 | 48.400 | 平均 | 38.17 | 46.21 | 40.21 | 28.10 | 45.600 | ||
| psbF | 40.83 | 47.50 | 45.00 | 30.00 | 61.000 | Mean | |||||||
Table 4 GC content and ENC of 48 CDS codons from the chloroplast of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ in Jiangxi Yanshan
| 基因 Gene | GC含量 GC content/% | 基因 Gene | GC含量 GC content/% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 平均 GCall | 第一密码子GC1 | 第二密码子GC2 | 第三密码子GC3 | 有效密码子数ENC | 平均 GCall | 第一密码子GC1 | 第二密码子GC2 | 第三密码子GC3 | 有效密码子数ENC | ||||
| accD | 34.62 | 38.62 | 37.40 | 27.85 | 44.206 | psbH | 42.34 | 48.65 | 47.30 | 31.08 | 43.164 | ||
| atpA | 40.03 | 55.71 | 39.17 | 25.20 | 44.586 | psbI | 36.94 | 48.65 | 29.73 | 32.43 | 40.922 | ||
| atpB | 42.48 | 56.91 | 41.68 | 28.86 | 48.495 | psbJ | 41.46 | 41.46 | 53.66 | 29.27 | 20.769 | ||
| atpE | 41.67 | 52.94 | 41.91 | 30.15 | 46.888 | psbK | 37.57 | 42.86 | 36.51 | 33.33 | 42.160 | ||
| atpF | 37.73 | 50.00 | 35.16 | 28.02 | 39.808 | psbL | 29.06 | 35.90 | 28.21 | 23.08 | 50.514 | ||
| atpH | 44.31 | 62.20 | 48.78 | 21.95 | 49.183 | psbM | 28.57 | 42.86 | 25.71 | 17.14 | 31.500 | ||
| atpI | 38.04 | 49.19 | 37.90 | 27.02 | 43.152 | psbT | 35.29 | 41.18 | 35.29 | 29.41 | 34.772 | ||
| ccsA | 32.1 | 32.10 | 37.96 | 26.23 | 43.768 | psbZ | 34.39 | 38.10 | 42.86 | 22.22 | 51.110 | ||
| cemA | 31.45 | 39.62 | 27.04 | 27.67 | 40.639 | rbcL | 43.04 | 57.38 | 43.66 | 28.07 | 47.314 | ||
| clpP1 | 42.69 | 59.61 | 36.95 | 31.53 | 59.229 | rpl2 | 43.80 | 50.73 | 49.27 | 31.39 | 50.648 | ||
| matK | 31.58 | 38.99 | 31.19 | 24.56 | 46.593 | rpl2 | 43.80 | 50.73 | 49.27 | 31.39 | 50.648 | ||
| ndhA | 35.81 | 44.51 | 37.91 | 25.00 | 44.550 | rpl14 | 39.02 | 53.66 | 38.21 | 25.20 | 56.951 | ||
| ndhB | 37.77 | 42.07 | 39.53 | 31.70 | 47.366 | rpl16 | 43.87 | 50.74 | 53.68 | 27.21 | 40.180 | ||
| ndhB | 37.77 | 42.07 | 39.53 | 31.70 | 47.366 | rpl20 | 36.44 | 37.29 | 44.92 | 27.12 | 43.650 | ||
| ndhC | 37.47 | 49.59 | 33.88 | 28.93 | 45.634 | rpl22 | 37.63 | 45.16 | 40.32 | 27.42 | 51.065 | ||
| ndhD | 35.46 | 39.72 | 38.52 | 28.14 | 49.596 | rpl23 | 37.04 | 41.11 | 40.00 | 30.00 | 52.839 | ||
| ndhE | 31.37 | 38.24 | 33.33 | 22.55 | 42.469 | rpl23 | 37.04 | 41.11 | 40.00 | 30.00 | 52.839 | ||
| ndhF | 33.06 | 37.42 | 36.88 | 24.90 | 45.696 | rpl32 | 33.91 | 34.48 | 44.83 | 22.41 | 34.549 | ||
| ndhG | 33.15 | 42.37 | 33.33 | 23.73 | 43.846 | rpl33 | 34.33 | 38.81 | 38.81 | 25.37 | 42.754 | ||
| ndhH | 38.16 | 51.78 | 35.79 | 26.90 | 48.229 | rpl36 | 37.72 | 42.11 | 47.37 | 23.68 | 46.750 | ||
| ndhI | 34.44 | 43.89 | 36.67 | 22.78 | 44.213 | rpoA | 35.00 | 45.59 | 34.41 | 25.00 | 49.552 | ||
| ndhJ | 39.2 | 48.43 | 37.74 | 31.45 | 51.139 | rpoB | 39.05 | 50.23 | 37.74 | 29.17 | 49.169 | ||
| ndhK | 38.82 | 43.37 | 43.37 | 29.72 | 51.528 | rpoC1 | 38.40 | 50.29 | 38.16 | 26.75 | 49.009 | ||
| pafI | 39.45 | 47.34 | 40.83 | 30.18 | 55.939 | rpoC2 | 37.48 | 46.02 | 37.63 | 28.80 | 50.022 | ||
| pafII | 38.92 | 44.86 | 40.54 | 31.35 | 51.331 | rps2 | 38.96 | 43.88 | 43.04 | 29.96 | 45.542 | ||
| pbf1 | 46.21 | 52.27 | 43.18 | 43.18 | 48.475 | rps3 | 34.86 | 46.58 | 34.25 | 23.74 | 45.101 | ||
| petA | 40.5 | 53.58 | 35.51 | 32.4 | 51.521 | rps4 | 37.79 | 49.01 | 38.12 | 26.24 | 50.288 | ||
| petB | 39.66 | 47.22 | 41.67 | 30.09 | 42.013 | rps7 | 39.10 | 52.56 | 44.87 | 19.87 | 44.746 | ||
| petD | 38.72 | 50.93 | 39.13 | 26.09 | 44.396 | rps7 | 39.10 | 52.56 | 44.87 | 19.87 | 44.746 | ||
| petG | 38.6 | 55.26 | 31.58 | 28.95 | 33.100 | rps8 | 36.09 | 41.35 | 42.11 | 24.81 | 42.540 | ||
| petL | 37.5 | 37.50 | 46.88 | 28.12 | 35.182 | rps11 | 44.12 | 55.40 | 57.55 | 19.42 | 41.507 | ||
| petN | 41.11 | 50.00 | 43.33 | 30.00 | 30.698 | rps12 | 41.94 | 52.42 | 48.39 | 25.00 | 43.732 | ||
| psaA | 43.01 | 51.93 | 43.68 | 33.42 | 49.161 | rps12 | 41.94 | 52.42 | 48.39 | 25.00 | 43.732 | ||
| psaB | 40.91 | 48.44 | 42.99 | 31.29 | 48.022 | rps14 | 41.58 | 42.57 | 46.53 | 35.64 | 37.716 | ||
| psaC | 43.9 | 43.90 | 53.66 | 34.15 | 48.358 | rps15 | 34.05 | 40.86 | 29.03 | 32.26 | 49.599 | ||
| psaI | 35.14 | 43.24 | 27.03 | 35.14 | 44.365 | rps16 | 35.86 | 50.63 | 36.71 | 20.25 | 42.450 | ||
| psaJ | 35.66 | 41.86 | 39.53 | 25.58 | 42.629 | rps18 | 34.31 | 35.29 | 44.12 | 23.53 | 37.936 | ||
| psbA | 42.66 | 49.72 | 43.79 | 34.46 | 41.107 | rps19 | 35.84 | 43.01 | 39.78 | 24.73 | 42.905 | ||
| psbB | 43.42 | 54.42 | 46.37 | 29.47 | 48.296 | ycf1 | 30.85 | 37.27 | 30.03 | 25.25 | 47.451 | ||
| psbC | 44.37 | 54.33 | 46.54 | 32.25 | 45.499 | ycf2 | 37.43 | 41.19 | 34.60 | 36.51 | 52.960 | ||
| psbD | 42.18 | 51.98 | 43.50 | 31.07 | 44.142 | ycf2 | 37.43 | 41.19 | 34.60 | 36.51 | 52.960 | ||
| psbE | 40.08 | 42.86 | 46.43 | 30.95 | 48.400 | 平均 | 38.17 | 46.21 | 40.21 | 28.10 | 45.600 | ||
| psbF | 40.83 | 47.50 | 45.00 | 30.00 | 61.000 | Mean | |||||||
| 氨基酸 Amino acid | 密码子 Codon | 数量 Amount | 相对使用度RSCU | 氨基酸 Amino acid | 密码子 Codon | 数量 Amount | 相对使用度RSCU | ||
|---|---|---|---|---|---|---|---|---|---|
| 丙氨酸 Ala | GCA | 420 | 1.187 28 | 赖氨酸 Lys | AAA | 1 036 | 1.48 318 | ||
| GCC | 215 | 0.607 774 | AAG | 361 | 0.516 822 | ||||
| GCG | 145 | 0.409 894 | 蛋氨酸Met | AUG | 596 | 1.993 31 | |||
| GCU | 635 | 1.795 05 | GUG | 2 | 0.006 688 96 | ||||
| 精氨酸 Arg | AGA | 521 | 1.953 75 | 苯丙氨酸Phe | UUC | 536 | 0.724 324 | ||
| AGG | 154 | 0.577 5 | UUU | 944 | 1.275 68 | ||||
| CGA | 358 | 1.342 5 | 脯氨酸 Pro | CCA | 322 | 1.194 81 | |||
| CGC | 96 | 0.36 | CCC | 197 | 0.730 983 | ||||
| CGG | 113 | 0.423 75 | CCG | 123 | 0.456 401 | ||||
| CGU | 358 | 1.342 5 | CCU | 436 | 1.617 81 | ||||
| 天冬酰胺 Asn | AAC | 290 | 0.464 | 丝氨酸 Ser | AGC | 104 | 0.298 851 | ||
| AAU | 960 | 1.536 | AGU | 447 | 1.284 48 | ||||
| 天冬氨酸 Asp | GAC | 213 | 0.394 81 | UCA | 444 | 1.275 86 | |||
| GAU | 866 | 1.605 19 | UCC | 339 | 0.974 138 | ||||
| 半胱氨酸 Cys | UGC | 74 | 0.485 246 | UCG | 179 | 0.514 368 | |||
| UGU | 231 | 1.514 75 | UCU | 575 | 1.652 3 | ||||
| 谷氨酰胺 Gln | CAA | 690 | 1.490 28 | 终止密码子Ter | UAA | 38 | 1.357 14 | ||
| CAG | 236 | 0.509 719 | UAG | 26 | 0.928 571 | ||||
| 谷氨酸 Glu | GAA | 1 035 | 1.483 87 | UGA | 20 | 0.714 286 | |||
| GAG | 360 | 0.516 129 | 苏氨酸 Thr | ACA | 433 | 1.277 29 | |||
| 甘氨酸 Gly | GGA | 719 | 1.632 24 | ACC | 247 | 0.728 614 | |||
| GGC | 161 | 0.365 494 | ACG | 149 | 0.439 528 | ||||
| GGG | 305 | 0.692 395 | ACU | 527 | 1.554 57 | ||||
| GGU | 577 | 1.309 88 | 色氨酸Trp | UGG | 443 | 1 | |||
| 组氨酸 His | CAC | 153 | 0.485 714 | 酪氨酸 Tyr | UAC | 208 | 0.428 424 | ||
| CAU | 477 | 1.514 29 | UAU | 763 | 1.571 58 | ||||
| 异亮氨酸 Ile | AUA | 702 | 0.950 79 | 缬氨酸 Val | GUA | 542 | 1.503 47 | ||
| AUC | 424 | 0.574 266 | GUC | 186 | 0.515 95 | ||||
| AUU | 1 089 | 1.474 94 | GUG | 194 | 0.538 141 | ||||
| 亮氨酸 Leu | CUA | 358 | 0.792 62 | GUU | 520 | 1.442 44 | |||
| CUC | 186 | 0.411 808 | |||||||
| CUG | 163 | 0.360 886 | |||||||
| CUU | 584 | 1.292 99 | |||||||
| UUA | 834 | 1.846 49 | |||||||
| UUG | 585 | 1.295 2 |
Table 5 RSCU analysis of amino acid codons in the chloroplast genome of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
| 氨基酸 Amino acid | 密码子 Codon | 数量 Amount | 相对使用度RSCU | 氨基酸 Amino acid | 密码子 Codon | 数量 Amount | 相对使用度RSCU | ||
|---|---|---|---|---|---|---|---|---|---|
| 丙氨酸 Ala | GCA | 420 | 1.187 28 | 赖氨酸 Lys | AAA | 1 036 | 1.48 318 | ||
| GCC | 215 | 0.607 774 | AAG | 361 | 0.516 822 | ||||
| GCG | 145 | 0.409 894 | 蛋氨酸Met | AUG | 596 | 1.993 31 | |||
| GCU | 635 | 1.795 05 | GUG | 2 | 0.006 688 96 | ||||
| 精氨酸 Arg | AGA | 521 | 1.953 75 | 苯丙氨酸Phe | UUC | 536 | 0.724 324 | ||
| AGG | 154 | 0.577 5 | UUU | 944 | 1.275 68 | ||||
| CGA | 358 | 1.342 5 | 脯氨酸 Pro | CCA | 322 | 1.194 81 | |||
| CGC | 96 | 0.36 | CCC | 197 | 0.730 983 | ||||
| CGG | 113 | 0.423 75 | CCG | 123 | 0.456 401 | ||||
| CGU | 358 | 1.342 5 | CCU | 436 | 1.617 81 | ||||
| 天冬酰胺 Asn | AAC | 290 | 0.464 | 丝氨酸 Ser | AGC | 104 | 0.298 851 | ||
| AAU | 960 | 1.536 | AGU | 447 | 1.284 48 | ||||
| 天冬氨酸 Asp | GAC | 213 | 0.394 81 | UCA | 444 | 1.275 86 | |||
| GAU | 866 | 1.605 19 | UCC | 339 | 0.974 138 | ||||
| 半胱氨酸 Cys | UGC | 74 | 0.485 246 | UCG | 179 | 0.514 368 | |||
| UGU | 231 | 1.514 75 | UCU | 575 | 1.652 3 | ||||
| 谷氨酰胺 Gln | CAA | 690 | 1.490 28 | 终止密码子Ter | UAA | 38 | 1.357 14 | ||
| CAG | 236 | 0.509 719 | UAG | 26 | 0.928 571 | ||||
| 谷氨酸 Glu | GAA | 1 035 | 1.483 87 | UGA | 20 | 0.714 286 | |||
| GAG | 360 | 0.516 129 | 苏氨酸 Thr | ACA | 433 | 1.277 29 | |||
| 甘氨酸 Gly | GGA | 719 | 1.632 24 | ACC | 247 | 0.728 614 | |||
| GGC | 161 | 0.365 494 | ACG | 149 | 0.439 528 | ||||
| GGG | 305 | 0.692 395 | ACU | 527 | 1.554 57 | ||||
| GGU | 577 | 1.309 88 | 色氨酸Trp | UGG | 443 | 1 | |||
| 组氨酸 His | CAC | 153 | 0.485 714 | 酪氨酸 Tyr | UAC | 208 | 0.428 424 | ||
| CAU | 477 | 1.514 29 | UAU | 763 | 1.571 58 | ||||
| 异亮氨酸 Ile | AUA | 702 | 0.950 79 | 缬氨酸 Val | GUA | 542 | 1.503 47 | ||
| AUC | 424 | 0.574 266 | GUC | 186 | 0.515 95 | ||||
| AUU | 1 089 | 1.474 94 | GUG | 194 | 0.538 141 | ||||
| 亮氨酸 Leu | CUA | 358 | 0.792 62 | GUU | 520 | 1.442 44 | |||
| CUC | 186 | 0.411 808 | |||||||
| CUG | 163 | 0.360 886 | |||||||
| CUU | 584 | 1.292 99 | |||||||
| UUA | 834 | 1.846 49 | |||||||
| UUG | 585 | 1.295 2 |
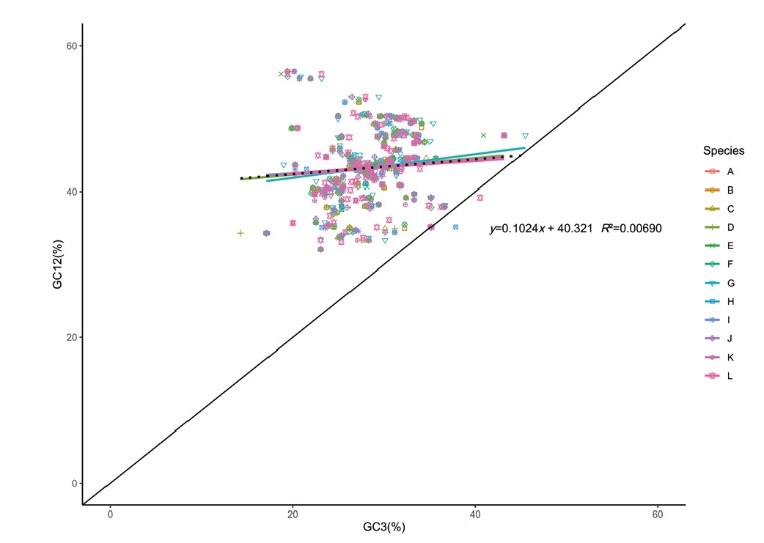
Fig. 7 Neutrality plot analysis of chloroplast genes of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan and its 11 related species A-L successively represents 11 related species(C. esculenta JN105690, C. esculenta NC_016753, R. vivipara MT884912, S. sp. 0020020418 MT884849, S. colocasiifolia NC_051952, A. peltata MT884904, A. fornicata MK636779, A. navicularis MN046882, L. gigantea NC_060475, E. hypnosum MT884866, A. cucullata NC_060696). The same below
| 氨基酸 Amino acid | 密码子 Codon | 低表达基因 Low expressed gene | 高表达基因 High expressed gene | 相对使用度差值 △RSCU | |||
|---|---|---|---|---|---|---|---|
| 相对使用度 RSCU | 数量 Amount | 相对使用度 RSCU | 数量 Amount | ||||
| Ala | GCU | 1.854 3 | 70 | 2 | 8 | 0.145 7 | |
| Arg | CGU | 0.672 414 | 39 | 1.875 | 3 | 1.202 586 | |
| AGG | 0.879 31 | 51 | 1.25 | 2 | 0.370 69 | ||
| CGC | 0.379 31 | 22 | 0.625 | 1 | 0.245 69 | ||
| Asn | AAC | 0.520 376 | 83 | 1 | 3 | 0.479 624 | |
| Gln | CAA | 1.299 49 | 128 | 1.5 | 3 | 0.200 51 | |
| Gly | GGU | 0.967 442 | 52 | 1.312 5 | 7 | 0.345 058 | |
| Ile | AUU | 1.334 93 | 186 | 2 | 12 | 0.665 07 | |
| Leu | UUA | 1.043 48 | 92 | 1.555 56 | 7 | 0.512 08 | |
| CUU | 1.417 77 | 125 | 1.555 56 | 7 | 0.137 79 | ||
| CUA | 0.793 951 | 70 | 1.111 11 | 5 | 0.317 159 | ||
| Lys | AAA | 1.254 78 | 197 | 1.538 46 | 10 | 0.283 68 | |
| Phe | UUU | 1.032 84 | 173 | 1.666 67 | 10 | 0.633 83 | |
| Pro | CCU | 1.301 44 | 68 | 1.6 | 4 | 0.298 56 | |
| Ser | AGU | 0.928 433 | 80 | 1.263 16 | 6 | 0.334 727 | |
| UCA | 1.334 62 | 115 | 1.473 68 | 7 | 0.139 06 | ||
| Ter | UGA | 0 | 0 | 0.6 | 1 | 0.6 | |
| UAG | 0.666 667 | 2 | 1.2 | 2 | 0.533 333 | ||
| Thr | ACU | 1.192 66 | 65 | 1.384 62 | 9 | 0.191 96 | |
| Tyr | UAC | 0.402 174 | 37 | 0.666 667 | 2 | 0.264 493 | |
| Val | GUA | 1.106 8 | 57 | 2.117 65 | 9 | 1.010 85 | |
Table 6 Putative optimal codons in chloroplast genome of C. esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan
| 氨基酸 Amino acid | 密码子 Codon | 低表达基因 Low expressed gene | 高表达基因 High expressed gene | 相对使用度差值 △RSCU | |||
|---|---|---|---|---|---|---|---|
| 相对使用度 RSCU | 数量 Amount | 相对使用度 RSCU | 数量 Amount | ||||
| Ala | GCU | 1.854 3 | 70 | 2 | 8 | 0.145 7 | |
| Arg | CGU | 0.672 414 | 39 | 1.875 | 3 | 1.202 586 | |
| AGG | 0.879 31 | 51 | 1.25 | 2 | 0.370 69 | ||
| CGC | 0.379 31 | 22 | 0.625 | 1 | 0.245 69 | ||
| Asn | AAC | 0.520 376 | 83 | 1 | 3 | 0.479 624 | |
| Gln | CAA | 1.299 49 | 128 | 1.5 | 3 | 0.200 51 | |
| Gly | GGU | 0.967 442 | 52 | 1.312 5 | 7 | 0.345 058 | |
| Ile | AUU | 1.334 93 | 186 | 2 | 12 | 0.665 07 | |
| Leu | UUA | 1.043 48 | 92 | 1.555 56 | 7 | 0.512 08 | |
| CUU | 1.417 77 | 125 | 1.555 56 | 7 | 0.137 79 | ||
| CUA | 0.793 951 | 70 | 1.111 11 | 5 | 0.317 159 | ||
| Lys | AAA | 1.254 78 | 197 | 1.538 46 | 10 | 0.283 68 | |
| Phe | UUU | 1.032 84 | 173 | 1.666 67 | 10 | 0.633 83 | |
| Pro | CCU | 1.301 44 | 68 | 1.6 | 4 | 0.298 56 | |
| Ser | AGU | 0.928 433 | 80 | 1.263 16 | 6 | 0.334 727 | |
| UCA | 1.334 62 | 115 | 1.473 68 | 7 | 0.139 06 | ||
| Ter | UGA | 0 | 0 | 0.6 | 1 | 0.6 | |
| UAG | 0.666 667 | 2 | 1.2 | 2 | 0.533 333 | ||
| Thr | ACU | 1.192 66 | 65 | 1.384 62 | 9 | 0.191 96 | |
| Tyr | UAC | 0.402 174 | 37 | 0.666 667 | 2 | 0.264 493 | |
| Val | GUA | 1.106 8 | 57 | 2.117 65 | 9 | 1.010 85 | |
| [1] | 刘星月, 朱强龙, 李慧英, 等. 红芽芋脱毒试管芋诱导及植株再生[J]. 园艺学报, 2020, 47(12): 2427-2438. |
|
Liu XY, Zhu QL, Li HY, et al. Induction and plant regeneration of virus-free microtuber in red bud taro[J]. Acta Hortic Sin, 2020, 47(12): 2427-2438.
doi: 10.16420/j.issn.0513-353x.2020-0470 |
|
| [2] | 李云, 牛丽亚, 涂瑾, 等. 亲水胶体对红芽芋全粉理化特性和消化特性的影响[J]. 中国粮油学报, 2020, 35(2): 12-17. |
| Li Y, Niu LY, Tu J, et al. Effect of hydrocolloids on physicochemical properties and digestive properties of red bud taro flour[J]. J Chin Cereals Oils Assoc, 2020, 35(2): 12-17. | |
| [3] | 邓接楼, 曹昊玮, 李玲, 等. 脱毒红芽芋试管芋盆栽种植的农艺性状分析[J]. 分子植物育种, 2019, 17(22): 7500-7506. |
| Deng JL, Cao HW, Li L, et al. Analysis on agronomic traits of virus-free test-tube taro planted in pots[J]. Mol Plant Breed, 2019, 17(22): 7500-7506. | |
| [4] | 肖慈生, 方加军, 肖月土, 等. 铅山红芽芋早熟栽培技术规程[J]. 长江蔬菜, 2018(15): 29-30. |
| Xiao CS, Fang JJ, Xiao YT, et al. Technical regulations for early maturing cultivation of Yanshan red bud taro[J]. J Chang Veg, 2018(15): 29-30. | |
| [5] | 余志平, 林海红, 余俊红, 等. 铅山红芽芋产业发展概况[J]. 长江蔬菜, 2016(20): 34-36. |
| Yu ZP, Lin HH, Yu JH, et al. Development of red bud taro industry in Yanshan[J]. J Chang Veg, 2016(20): 34-36. | |
| [6] |
姜绍通, 程元珍, 郑志, 等. 红芽芋营养成分分析及评价[J]. 食品科学, 2012, 33(11): 269-272.
doi: 10.7506/spkx1002-6630-201211057 |
| Jiang ST, Cheng YZ, Zheng Z, et al. Analysis and evaluation of nutritional components of red bud taro(Colocasia esulenla L. schott)[J]. Food Sci, 2012, 33(11): 269-272. | |
| [7] |
Wang QR, Huang ZR, Gao CS, et al. The complete chloroplast genome sequence of Rubus hirsutus Thunb. and a comparative analysis within Rubus species[J]. Genetica, 2021, 149(5/6): 299-311.
doi: 10.1007/s10709-021-00131-9 |
| [8] |
Zhao ZY, Wang X, Yu Y, et al. Complete chloroplast genome sequences of Dioscorea: Characterization, genomic resources, and phylogenetic analyses[J]. PeerJ, 2018, 6: e6032.
doi: 10.7717/peerj.6032 URL |
| [9] |
Daniell H, Lin CS, Yu M, et al. Chloroplast genomes: diversity, evolution, and applications in genetic engineering[J]. Genome Biol, 2016, 17(1): 134.
doi: 10.1186/s13059-016-1004-2 pmid: 27339192 |
| [10] |
Ahmed I, Matthews PJ, Biggs PJ, et al. Identification of chloroplast genome loci suitable for high-resolution phylogeographic studies of Colocasia esculenta(L.)Schott(Araceae)and closely related taxa[J]. Mol Ecol Resour, 2013, 13(5): 929-937.
doi: 10.1111/men.2013.13.issue-5 URL |
| [11] | Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data[J]. Nucleic Acids Res, 2017, 45(4): e18. |
| [12] |
Tillich M, Lehwark P, Pellizzer T, et al. GeSeq - versatile and accurate annotation of organelle genomes[J]. Nucleic Acids Res, 2017, 45(W1): W6-W11.
doi: 10.1093/nar/gkx391 URL |
| [13] |
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence[J]. Nucleic Acids Res, 1997, 25(5): 955-964.
doi: 10.1093/nar/25.5.955 pmid: 9023104 |
| [14] |
Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW(OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes[J]. Curr Genet, 2007, 52(5/6): 267-274.
doi: 10.1007/s00294-007-0161-y URL |
| [15] |
Thiel T, Michalek W, Varshney RK, et al. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley(Hordeum vulgare L.)[J]. Theor Appl Genet, 2003, 106(3): 411-422.
doi: 10.1007/s00122-002-1031-0 pmid: 12589540 |
| [16] |
Low SL, Yu CC, Ooi IH, et al. Extensive Miocene speciation in and out of Indochina: the biogeographic history of Typhonium sensu stricto(Araceae)and its implication for the assembly of Indochina flora[J]. J Syst Evol, 2021, 59(3): 419-428.
doi: 10.1111/jse.v59.3 URL |
| [17] |
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability[J]. Mol Biol Evol, 2013, 30(4): 772-780.
doi: 10.1093/molbev/mst010 pmid: 23329690 |
| [18] |
Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments[J]. PLoS One, 2010, 5(3): e9490.
doi: 10.1371/journal.pone.0009490 URL |
| [19] |
Sun LY, Jiang Z, Wan XX, et al. The complete chloroplast genome of Magnolia polytepala: comparative analyses offer implication for genetics and phylogeny of Yulania[J]. Gene, 2020, 736: 144410.
doi: 10.1016/j.gene.2020.144410 URL |
| [20] |
Ahmed I, Biggs PJ, Matthews PJ, et al. Mutational dynamics of aroid chloroplast genomes[J]. Genome Biol Evol, 2012, 4(12): 1316-1323.
doi: 10.1093/gbe/evs110 pmid: 23204304 |
| [21] |
Gao LQ, Li YL, Guo CC, et al. Indocalamus chongzhouensis(Poaceae: Bambusoideae), a new synonym of I. emeiensis: evidence from morphology and complete chloroplast genome data[J]. Phytotaxa, 2022, 542(1): 53-63.
doi: 10.11646/phytotaxa.542.1 URL |
| [22] |
Romero H, Zavala A, Musto H. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces[J]. Nucleic Acids Res, 2000, 28(10): 2084-2090.
doi: 10.1093/nar/28.10.2084 pmid: 10773076 |
| [23] |
Carlini DB, Chen Y, Stephan W. The relationship between third-codon position nucleotide content, codon bias, mRNA secondary structure and gene expression in the drosophilid alcohol dehydrogenase genes Adh and Adhr[J]. Genetics, 2001, 159(2): 623-633.
doi: 10.1093/genetics/159.2.623 pmid: 11606539 |
| [24] |
Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis[J]. Proc Natl Acad Sci USA, 1999, 96(8): 4482-4487.
doi: 10.1073/pnas.96.8.4482 pmid: 10200288 |
| [25] |
Sharp PM, Emery LR, Zeng K. Forces that influence the evolution of codon bias[J]. Philos Trans R Soc Lond B Biol Sci, 2010, 365(1544): 1203-1212.
doi: 10.1098/rstb.2009.0305 URL |
| [26] |
Takahashi D, Sakaguchi S, Isagi Y, et al. Comparative chloroplast genomics of series Sakawanum in genus Asarum(Aristolochiaceae)to develop single nucleotide polymorphisms(SNPs)and simple sequence repeat(SSR)markers[J]. J For Res, 2018, 23(6): 387-392.
doi: 10.1080/13416979.2018.1518649 URL |
| [27] |
Yamane K, Kawahara T. Size homoplasy and mutational behavior of chloroplast simple sequence repeats(cpSSRs)inferred from intra- and interspecific variations in four chloroplast regions of diploid and polyploid Triticum and Aegilops species[J]. Genet Resour Crop Evol, 2018, 65(3): 727-743.
doi: 10.1007/s10722-017-0567-4 URL |
| [28] |
Dugas DV, Hernandez D, Koenen EJM, et al. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP[J]. Sci Rep, 2015, 5: 16958.
doi: 10.1038/srep16958 pmid: 26592928 |
| [29] |
Park S, An B, Park S. Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation[J]. Sci Rep, 2018, 8(1): 13568.
doi: 10.1038/s41598-018-31938-w |
| [30] |
Raubeson LA, Peery R, Chumley TW, et al. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus[J]. BMC Genomics, 2007, 8: 174.
pmid: 17573971 |
| [31] | Li X, Li Y, Zang M, et al. Complete chloroplast genome sequence and phylogenetic analysis of Quercus acutissima[J]. Int J Mol Sci, 2018, 19(8): E2443. |
| [32] |
Gu C, Ma L, Wu Z, et al. Comparative analyses of chloroplast genomes from 22 Lythraceae species: inferences for phylogenetic relationships and genome evolution within Myrtales[J]. BMC Plant Biol, 2019, 19(1): 281.
doi: 10.1186/s12870-019-1870-3 pmid: 31242865 |
| [33] |
Dong WP, Xu C, Li CH, et al. ycf1, the most promising plastid DNA barcode of land plants[J]. Sci Rep, 2015, 5: 8348.
doi: 10.1038/srep08348 pmid: 25672218 |
| [34] |
Amar MH. ycf1-ndhF genes, the most promising plastid genomic barcode, sheds light on phylogeny at low taxonomic levels in Prunus persica[J]. J Genet Eng Biotechnol, 2020, 18(1): 42.
doi: 10.1186/s43141-020-00057-3 |
| [1] | ZHU Bin, GAN Chen-chen, WANG Hong-cheng. Characteristics of the Complete Chloroplast Genome of Dendrobium thyrsiflorum and Its Phylogenetic Relationship Analysis [J]. Biotechnology Bulletin, 2021, 37(5): 38-47. |
| [2] | CHEN Wei-min, CHAI Hong-mei, YANG Wei-xian, MA Yuan-hao, ZHAO Yong-chang. Characterization of B Mating Loci Identification and Application in Cyclocybe aegerita and C. salicacola [J]. Biotechnology Bulletin, 2021, 37(11): 57-64. |
| [3] | ZHANG Guang-zhi, WANG Jia-ning, WU Xiao-qing, ZHOU Fang-yuan, ZHANG Xin-jian, ZHAO Xiao-yan, XIE Xue-ying, ZHOU Hong-zi. Diversity and Functional Activity of Trichoderma in the Rhizosphere Soil from Facility Tomato Production [J]. Biotechnology Bulletin, 2018, 34(4): 179-185. |
| [4] | LIANG Han-qiao, Dilidaer Kudereti, BAI Fei-rong, MIAO Lei, LI Nan-nan, LIU Su-hui, CHEN Jian-guo, LIU Yang, SUN Yu-ping. Isolation and Identification of Endophytic Fungi from the Rhizomes of Xinjiang-specific Plant Brassica rapa [J]. Biotechnology Bulletin, 2017, 33(2): 131-136. |
| [5] | CAI Dong-mei, GONG Guo-li. The Current Status and Future Perspectives of Production of Biopharmaceuticals in Escherichia coli [J]. Biotechnology Bulletin, 2016, 32(8): 34-40. |
| [6] | HU Wen-zhe, TAN Ze-wen, WANG Yong, XU Xian-wei, TAN Zhi-yuan. Isolation,Identification and Phylogenetic Analysis of Endophytic Diazotrophs in Tengxian Medical-use Oryza officinalis [J]. Biotechnology Bulletin, 2016, 32(6): 111-119. |
| [7] | XIAO Jing,ZHA Xing-qin,CHENG Wen-min,HUO Jin-long,PAN Wei-rong,WANG Shu-yan,WANG Pei,LIU Hai-jing,ZHANG Xiu-qiong,ZENG Yang-zhi. Cloning,Sequence and Phylogenetic Analysis of CATSPER3 Gene in Banna Mini-pig Inbred Line [J]. Biotechnology Bulletin, 2016, 32(12): 103-107. |
| [8] | Chen Chunlin, Liu Xiang, Ju Xiong. Expression and Purification of Outer Membrane Lipoprotein OprF in Recombinant Pseudomonas aeruginosa,and Preparation and Identification of Polyclonal Antibody [J]. Biotechnology Bulletin, 2015, 31(8): 213-218. |
| [9] | Ni Yujia,Zhou Minyu,Ouyang Jia,Zheng Zhaojuan,Yong Qiang. Cloning,Expression of Thermostable β-mannanase and the Preparation of Mannooligosaccharide [J]. Biotechnology Bulletin, 2014, 0(6): 181-186. |
| [10] | Wang Fei, Liu Kang, Fan Jiatong, Cheng Shouliang, Wang Ziyuan. Isolation and Identification of a Halotolerant—Vibrio sp. K1-L [J]. Biotechnology Bulletin, 2014, 0(4): 127-131. |
| [11] | Hou Xindong Yin Shuai Sheng Guilian Cheng Dandan Lai Xulong. Study on Phylogenetic Relationships of Ten Urticaceae Species Based on nrDNA ITS Sequences [J]. Biotechnology Bulletin, 2013, 0(8): 68-73. |
| [12] | You Huan, Zhou Atao, Yue Liangliang, Cun Dongyi, Li Min, Ding Yuanming. Research on nrDNA ITS Pseudogenes in Plants [J]. Biotechnology Bulletin, 2013, 0(11): 14-18. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||