








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (6): 248-258.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1141
Previous Articles Next Articles
ZHANG Lu-yang1( ), HAN Wen-long1, XU Xiao-wen2, YAO Jian2, LI Fang-fang3, TIAN Xiao-yuan3, ZHANG Zhi-qiang1(
), HAN Wen-long1, XU Xiao-wen2, YAO Jian2, LI Fang-fang3, TIAN Xiao-yuan3, ZHANG Zhi-qiang1( )
)
Received:2022-09-16
Online:2023-06-26
Published:2023-07-07
Contact:
ZHANG Zhi-qiang
E-mail:zhangluyang97@126.com;xiao_qiang8866@163.com
ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family[J]. Biotechnology Bulletin, 2023, 39(6): 248-258.
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| NtTCP1 | TGTCGACGTCGGTAGAAGCTA | CGTCCATCGACTTTGCTATGA |
| NtTCP2 | CAGCCACTCCCTATGTCATCA | CATGGCCATTAGTTCCCAAAT |
| NtTCP3 | TATGCAGTTCCAGAGCTTCCA | TTTGGTGGTGCAGCAAAATAG |
| NtTCP4 | ATAGCAGCCACTGGAACTGGT | TTGAAACAAACCTCGTCGTTG |
| NtTCP5 | TGGGTCACAAGTCTGAAGGTG | GGGATTGTACCTGTTCCCGTA |
| NtTCP6 | AACCTTCTAAAGACCGCCACA | TTGTGACCCAATTCTCTGGTG |
| NtTCP7 | ACGCGAATTGGGTCATAAATC | GGAATTGTTCCATTGCCTGTT |
| NtTCP8 | GGGCAGTGGGCTCTAGTAATG | CCACAACTGAGGTGGATACGA |
| NtTCP9 | CCATTGCATTTCATGCCTAGA | GACATTCCCAACCCAAGATGT |
| NtTCP10 | TGACGGTAGAGGAAGGCGTAT | TTTGGCCATCAGACTTGTGAC |
| NtTCP11 | ATAGCAGCCACTGGAACTGGT | TGGGAGAGCGAAAAATTAGGA |
| NtTCP12 | GCAATGATAATGCTGGGGAAT | GCATTTGCATTGTCACCACTT |
| NtTCP13 | TTTACCCCACTTGGGTTTGAC | AACAAGTACCCACCACCTTGG |
| NtTCP14 | TGTCGACGTCGGTAGAAGCTA | CCATTATTGCCAGCATCAGAA |
| NtTCP15 | ATTTGCGAGGAGGCACTATGT | TCAACTTTCGTGTGCCTGTCT |
| NtTCP16 | TTGCTTCATCACCAAAACCAG | GACCGAGTTCTACTGGCTGCT |
| NtTCP17 | GTTGAAGGCAGAGGAAGGAGA | TGTTGCAGCAATTATGGAAGG |
| NtTCP18 | CCTCCGAAGAAACAAACCAAG | GTTTCTCCGTCGGACTTATGG |
| NtTCP19 | TCTGGATGTTACCGGTGAGTG | ACCATTGAACCCAACTGAACC |
| NtTCP20 | TGACGGTAGAGGAAGGCGTAT | TTTGACCATCGGACTTGTGAC |
Table 1 Real-time PCR primers for tobacco TCP gene expression analysis
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| NtTCP1 | TGTCGACGTCGGTAGAAGCTA | CGTCCATCGACTTTGCTATGA |
| NtTCP2 | CAGCCACTCCCTATGTCATCA | CATGGCCATTAGTTCCCAAAT |
| NtTCP3 | TATGCAGTTCCAGAGCTTCCA | TTTGGTGGTGCAGCAAAATAG |
| NtTCP4 | ATAGCAGCCACTGGAACTGGT | TTGAAACAAACCTCGTCGTTG |
| NtTCP5 | TGGGTCACAAGTCTGAAGGTG | GGGATTGTACCTGTTCCCGTA |
| NtTCP6 | AACCTTCTAAAGACCGCCACA | TTGTGACCCAATTCTCTGGTG |
| NtTCP7 | ACGCGAATTGGGTCATAAATC | GGAATTGTTCCATTGCCTGTT |
| NtTCP8 | GGGCAGTGGGCTCTAGTAATG | CCACAACTGAGGTGGATACGA |
| NtTCP9 | CCATTGCATTTCATGCCTAGA | GACATTCCCAACCCAAGATGT |
| NtTCP10 | TGACGGTAGAGGAAGGCGTAT | TTTGGCCATCAGACTTGTGAC |
| NtTCP11 | ATAGCAGCCACTGGAACTGGT | TGGGAGAGCGAAAAATTAGGA |
| NtTCP12 | GCAATGATAATGCTGGGGAAT | GCATTTGCATTGTCACCACTT |
| NtTCP13 | TTTACCCCACTTGGGTTTGAC | AACAAGTACCCACCACCTTGG |
| NtTCP14 | TGTCGACGTCGGTAGAAGCTA | CCATTATTGCCAGCATCAGAA |
| NtTCP15 | ATTTGCGAGGAGGCACTATGT | TCAACTTTCGTGTGCCTGTCT |
| NtTCP16 | TTGCTTCATCACCAAAACCAG | GACCGAGTTCTACTGGCTGCT |
| NtTCP17 | GTTGAAGGCAGAGGAAGGAGA | TGTTGCAGCAATTATGGAAGG |
| NtTCP18 | CCTCCGAAGAAACAAACCAAG | GTTTCTCCGTCGGACTTATGG |
| NtTCP19 | TCTGGATGTTACCGGTGAGTG | ACCATTGAACCCAACTGAACC |
| NtTCP20 | TGACGGTAGAGGAAGGCGTAT | TTTGACCATCGGACTTGTGAC |
| 基因名称 Gene name | 基因组登录号 Gene ID | 氨基酸数 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 亲水性 GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| NtTCP1 | Nitab4.5_0000069g0220.1 | 247 | 25.895 | 7.93 | -0.442 | 细胞核 Nucleus |
| NtTCP2 | Nitab4.5_0000206g0010.1 | 442 | 48.502 | 8.55 | -0.907 | 细胞核 Nucleus |
| NtTCP3 | Nitab4.5_0000246g0120.1 | 323 | 35.413 | 5.51 | -0.822 | 细胞核 Nucleus |
| NtTCP4 | Nitab4.5_0000269g0060.1 | 367 | 39.854 | 7.37 | -0.715 | 细胞核 Nucleus |
| NtTCP5 | Nitab4.5_0000303g0270.1 | 326 | 34.960 | 6.87 | -0.784 | 细胞核 Nucleus |
| NtTCP6 | Nitab4.5_0000365g0240.1 | 412 | 44.290 | 8.99 | -0.746 | 细胞核 Nucleus |
| NtTCP7 | Nitab4.5_0000519g0050.1 | 222 | 24.004 | 9.82 | -0.486 | 细胞核 Nucleus |
| NtTCP8 | Nitab4.5_0000832g0020.1 | 357 | 38.007 | 5.53 | -0.448 | 细胞核 Nucleus |
| NtTCP9 | Nitab4.5_0000948g0150.1 | 556 | 59.693 | 7.19 | -0.758 | 细胞核 Nucleus |
| NtTCP10 | Nitab4.5_0001089g0050.1 | 161 | 16.833 | 7.33 | -0.244 | 细胞核 Nucleus |
| NtTCP11 | Nitab4.5_0001188g0020.1 | 399 | 43.838 | 6.89 | -0.823 | 细胞核 Nucleus |
| NtTCP12 | Nitab4.5_0003077g0060.1 | 596 | 66.701 | 6.14 | -0.808 | 细胞核 Nucleus |
| NtTCP13 | Nitab4.5_0003202g0050.1 | 420 | 46.357 | 6.57 | -0.793 | 细胞核 Nucleus |
| NtTCP14 | Nitab4.5_0004301g0070.1 | 300 | 31.547 | 8.65 | -0.324 | 细胞核 Nucleus |
| NtTCP15 | Nitab4.5_0005379g0040.1 | 359 | 38.206 | 5.43 | -0.444 | 细胞核 Nucleus |
| NtTCP16 | Nitab4.5_0005709g0020.1 | 589 | 65.994 | 6.65 | -0.817 | 细胞核 Nucleus |
| NtTCP17 | Nitab4.5_0007375g0060.1 | 295 | 32.098 | 7.14 | -0.744 | 细胞核 Nucleus |
| NtTCP18 | Nitab4.5_0007913g0020.1 | 544 | 58.332 | 7.00 | -0.761 | 细胞核 Nucleus |
| NtTCP19 | Nitab4.5_0009198g0010.1 | 419 | 44.840 | 8.04 | -0.804 | 细胞核 Nucleus |
| NtTCP20 | Nitab4.5_0011777g0010.1 | 261 | 27.539 | 9.71 | -0.432 | 细胞核 Nucleus |
Table 2 Basic information of TCP gene family members in tobacco
| 基因名称 Gene name | 基因组登录号 Gene ID | 氨基酸数 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 亲水性 GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| NtTCP1 | Nitab4.5_0000069g0220.1 | 247 | 25.895 | 7.93 | -0.442 | 细胞核 Nucleus |
| NtTCP2 | Nitab4.5_0000206g0010.1 | 442 | 48.502 | 8.55 | -0.907 | 细胞核 Nucleus |
| NtTCP3 | Nitab4.5_0000246g0120.1 | 323 | 35.413 | 5.51 | -0.822 | 细胞核 Nucleus |
| NtTCP4 | Nitab4.5_0000269g0060.1 | 367 | 39.854 | 7.37 | -0.715 | 细胞核 Nucleus |
| NtTCP5 | Nitab4.5_0000303g0270.1 | 326 | 34.960 | 6.87 | -0.784 | 细胞核 Nucleus |
| NtTCP6 | Nitab4.5_0000365g0240.1 | 412 | 44.290 | 8.99 | -0.746 | 细胞核 Nucleus |
| NtTCP7 | Nitab4.5_0000519g0050.1 | 222 | 24.004 | 9.82 | -0.486 | 细胞核 Nucleus |
| NtTCP8 | Nitab4.5_0000832g0020.1 | 357 | 38.007 | 5.53 | -0.448 | 细胞核 Nucleus |
| NtTCP9 | Nitab4.5_0000948g0150.1 | 556 | 59.693 | 7.19 | -0.758 | 细胞核 Nucleus |
| NtTCP10 | Nitab4.5_0001089g0050.1 | 161 | 16.833 | 7.33 | -0.244 | 细胞核 Nucleus |
| NtTCP11 | Nitab4.5_0001188g0020.1 | 399 | 43.838 | 6.89 | -0.823 | 细胞核 Nucleus |
| NtTCP12 | Nitab4.5_0003077g0060.1 | 596 | 66.701 | 6.14 | -0.808 | 细胞核 Nucleus |
| NtTCP13 | Nitab4.5_0003202g0050.1 | 420 | 46.357 | 6.57 | -0.793 | 细胞核 Nucleus |
| NtTCP14 | Nitab4.5_0004301g0070.1 | 300 | 31.547 | 8.65 | -0.324 | 细胞核 Nucleus |
| NtTCP15 | Nitab4.5_0005379g0040.1 | 359 | 38.206 | 5.43 | -0.444 | 细胞核 Nucleus |
| NtTCP16 | Nitab4.5_0005709g0020.1 | 589 | 65.994 | 6.65 | -0.817 | 细胞核 Nucleus |
| NtTCP17 | Nitab4.5_0007375g0060.1 | 295 | 32.098 | 7.14 | -0.744 | 细胞核 Nucleus |
| NtTCP18 | Nitab4.5_0007913g0020.1 | 544 | 58.332 | 7.00 | -0.761 | 细胞核 Nucleus |
| NtTCP19 | Nitab4.5_0009198g0010.1 | 419 | 44.840 | 8.04 | -0.804 | 细胞核 Nucleus |
| NtTCP20 | Nitab4.5_0011777g0010.1 | 261 | 27.539 | 9.71 | -0.432 | 细胞核 Nucleus |
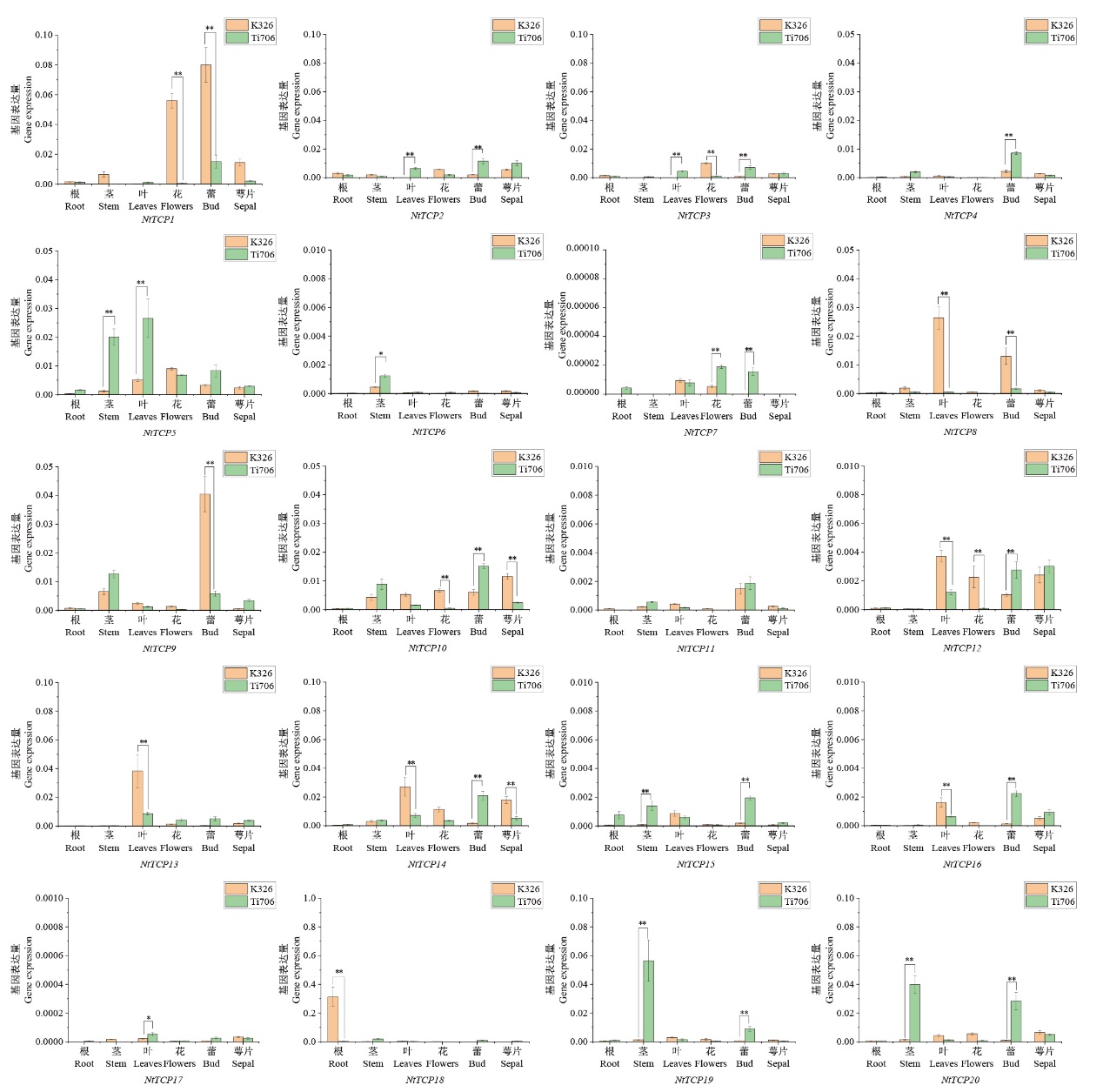
Fig. 6 Expressions of tobacco TCP gene family in the different tissues of K326 and Ti706 The data used are the mean ± SD of three biological replicates. * on the column indicates significant differences in P< 0.05 levels among different treatments, while ** indicates significant differences in P<0.01 levels among different treatments. The same below
| [1] | 贾中林, 尹启生, 戴华鑫, 等. 烤烟上部烟叶成熟生理特性研究进展[J]. 烟草科技, 2022, 55(7): 99-112. |
| Jia ZL, Yin QS, Dai HX, et al. Research progress on physiological characteristics of mature upper leaves of flue-cured tobacco[J]. Tob Sci & Technol, 2022, 55(7): 99-112. | |
| [2] |
Cubas P, Coen E, Zapater JM. Ancient asymmetries in the evolution of flowers[J]. Curr Biol, 2001, 11(13): 1050-1052.
pmid: 11470410 |
| [3] |
Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize[J]. Nature, 1997, 386(6624): 485-488.
doi: 10.1038/386485a0 |
| [4] |
Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene[J]. Plant Cell, 1997, 9(9): 1607-1619.
doi: 10.1105/tpc.9.9.1607 pmid: 9338963 |
| [5] |
Martín-Trillo M, Cubas P. TCP genes: a family snapshot ten years later[J]. Trends Plant Sci, 2010, 15(1): 31-39.
doi: 10.1016/j.tplants.2009.11.003 pmid: 19963426 |
| [6] |
Yao X, Ma H, Wang J, et al. Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa[J]. J Integr Plant Biol, 2007, 49(6): 885-897.
doi: 10.1111/jipb.2007.49.issue-6 URL |
| [7] | 关紫微, 曹希雅, 张先文, 等. 水稻TCP家族的全基因组鉴定及表达分析[J]. 分子植物育种, 2022(10):3145-3156. |
| Guan ZW, Cao XY, Zhang XW, et al. Genome-wide identification and expression analysis of TCP family in rice(Oryza sativa L.)[J]. Mol Plant Breed, 2022(10):3145-3156. | |
| [8] | 李菲, 何小红, 张宇斌, 等. 番茄TCP转录因子家族的鉴定和生物信息学分析[J]. 分子植物育种, 2018, 16(21): 6899-6906. |
| Li F, He XH, Zhang YB, et al. Identification and bioinformatics analysis of TCP transcription factor family in tomato[J]. Mol Plant Breed, 2018, 16(21): 6899-6906. | |
| [9] | 杨婷, 黎成, 申佳瑜, 等. 茄子TCP基因家族全基因组的鉴定与分析[J]. 生物工程学报, 2022(8):2974-2988. |
| Yang T, Li C, Shen JY, et al. Genome-wide identification and analysis of the TCP gene family in eggplant(Solanum melongena L.)[J]. Chin J Biotechnol, 2022(8):2974-2988. | |
| [10] | 冯建英, 李立芹, 李佳皓, 等. 马铃薯TCP家族转录因子鉴定与表达模式分析[J]. 基因组学与应用生物学, 2021, 40(S2): 2756-2764. |
| Feng JY, Li LQ, Li JH, et al. Identification and expression pattern analysis of TCP family transcription factors in potato[J]. Genom Appl Biol, 2021, 40(S2): 2756-2764. | |
| [11] | 刘京, 吕前前, 盛鸥, 等. 香蕉TCP全基因家族的鉴定及表达模式分析[J]. 分子植物育种, 2023, 21(5):1427-1437. |
| Liu J, Lu QQ, Sheng O, et al. Identification and expression pattern analysis of TCP gene family in banana[J]. Mol Plant Breed, 2023, 21(5):1427-1437. | |
| [12] |
An JX, Guo ZX, Gou XP, et al. TCP1 positively regulates the expression of DWF4 in Arabidopsis thaliana[J]. Plant Signal Behav, 2011, 6(8): 1117-1118.
doi: 10.4161/psb.6.8.15889 URL |
| [13] |
Guo ZX, Fujioka S, Blancaflor EB, et al. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana[J]. Plant Cell, 2010, 22(4): 1161-1173.
doi: 10.1105/tpc.109.069203 URL |
| [14] |
Nag A, King S, Jack T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis[J]. PNAS, 2009, 106(52): 22534-22539.
doi: 10.1073/pnas.0908718106 URL |
| [15] |
Resentini F, Felipo-Benavent A, Colombo L, et al. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana[J]. Mol Plant, 2015, 8(3): 482-485.
doi: 10.1016/j.molp.2014.11.018 URL |
| [16] |
Hervé C, Dabos P, Bardet C, et al. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development[J]. Plant Physiol, 2009, 149(3): 1462-1477.
doi: 10.1104/pp.108.126136 URL |
| [17] |
Aguilar-Martínez JA, Sinha N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development[J]. Front Plant Sci, 2013, 4: 406.
doi: 10.3389/fpls.2013.00406 pmid: 24137171 |
| [18] |
Palatnik JF, Allen E, Wu XL, et al. Control of leaf morphogenesis by microRNAs[J]. Nature, 2003, 425(6955): 257-263.
doi: 10.1038/nature01958 |
| [19] | 张雪莹, 尹一歌, 姜晶, 等. 参与番茄叶形发育的TCP转录因子的表达及生物信息分析[J]. 中国蔬菜, 2021(10): 45-56. |
| Zhang XY, Yin YG, Jiang J, et al. Expression of SlTCPs transcription factors involved in tomato leaf shape development and bioinformatic analysis[J]. China Veg, 2021(10): 45-56. | |
| [20] | 王景超. 植物TCP家族基因的研究进展[J]. 农业与技术, 2021, 41(18): 63-66. |
| Wang JC. Research progress of TCP family genes in plants[J]. Agric Technol, 2021, 41(18): 63-66. | |
| [21] |
Zhang X, Bao YL, Shan DQ, et al. Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice[J]. Plant Physiol, 2018, 177(1): 352-368.
doi: 10.1104/pp.17.01665 pmid: 29549093 |
| [22] |
Han X, Yu H, Yuan RR, et al. Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity[J]. iScience, 2019, 15: 611-622.
doi: 10.1016/j.isci.2019.04.005 URL |
| [23] |
Pruneda-Paz JL, Breton G, Para A, et al. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock[J]. Science, 2009, 323(5920): 1481-1485.
doi: 10.1126/science.1167206 pmid: 19286557 |
| [24] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [25] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [26] | 阮先乐, 张福丽, 王俊生. 番茄TCP基因家族全基因组鉴定和分析[J]. 基因组学与应用生物学, 2017, 36(6): 2539-2547. |
| Ruan XL, Zhang FL, Wang JS. Genome-wide identification and analysis of TCP gene family in tomato[J]. Genom Appl Biol, 2017, 36(6): 2539-2547. | |
| [27] | 周瑾, 李洁, 李晓燕, 等. 芹菜TCP基因家族全基因组鉴定、进化分析和表达研究[J]. 四川农业大学学报, 2022, 40(2): 145-155. |
| Zhou J, Li J, Li XY, et al. Genome-wide identification, evolution and expression analysis of TCP gene family in celery(Apium graveolens L.)[J]. J Sichuan Agric Univ, 2022, 40(2): 145-155. | |
| [28] | 王景超, 张君, 齐云, 等. 玉米TCP家族基因的表达分析[J]. 玉米科学, 2022, 30(1): 63-68. |
| Wang JC, Zhang J, Qi Y, et al. Expression analysis on TCP family genes of maize[J]. J Maize Sci, 2022, 30(1): 63-68. | |
| [29] |
Wang ST, Sun XL, Hoshino Y, et al. microRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice(Oryza sativa L.)[J]. PLoS One, 2014, 9(3): e91357.
doi: 10.1371/journal.pone.0091357 URL |
| [30] | 杨志晓, 王轶, 刘红峰, 等. 我国主栽烤烟品种亲缘关系及育种[J]. 中国烟草学报, 2013, 19(2): 34-41. |
| Yang ZX, Wang Y, Liu HF, et al. Genetic relationship in major flue-cured tobacco cultivars in China and its implication in variety breeding[J]. Acta Tabacaria Sin, 2013, 19(2): 34-41. | |
| [31] | 李坤杰, 谭杉杉, 孙勃, 等. 芥菜TCP转录因子家族全基因组鉴定及表达分析[J]. 四川农业大学学报, 2019, 37(4): 459-468. |
| Li KJ, Tan SS, Sun B, et al. Genome-wide identification and analysis of TCP transcription factor family in Brassica juncea[J]. J Sichuan Agric Univ, 2019, 37(4): 459-468. | |
| [32] |
Jiu ST, Xu Y, Wang JY, et al. Genome-wide identification, characterization, and transcript analysis of the TCP transcription factors in Vitis vinifera[J]. Front Genet, 2019, 10: 1276.
doi: 10.3389/fgene.2019.01276 URL |
| [1] | WU Qiao-yin, SHI You-zhi, LI Lin-lin, PENG Zheng, TAN Zai-yu, LIU Li-ping, ZHANG Juan, PAN Yong. In Situ Screening of Carotenoid Degrading Strains and the Application in Improving Quality and Aroma of Cigar [J]. Biotechnology Bulletin, 2023, 39(9): 192-201. |
| [2] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [3] | LIU Zhen-yin, DUAN Zhi-zhen, PENG Ting, WANG Tong-xin, WANG Jian. Establishment and Optimization of Virus-induced Gene Silencing System in Bougainvillea peruviana ‘Thimma’ [J]. Biotechnology Bulletin, 2023, 39(7): 123-130. |
| [4] | LI Wen-chen, LIU Xin, KANG Yue, LI Wei, QI Ze-zheng, YU Lu, WANG Fang. Optimization and Application of Tobacco Rattle Virus-induced Gene Silencing System in Soybean [J]. Biotechnology Bulletin, 2023, 39(7): 143-150. |
| [5] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [6] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [7] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [8] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [9] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [10] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [11] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [12] | SHEN Yun-xin, SHI Zhu-feng, ZHOU Xu-dong, LI Ming-gang, ZHANG Qing, FENG Lu-yao, CHEN Qi-bin, YANG Pei-wen. Isolation, Identification and Bio-activity of Three Bacillus Strains with Biocontrol Function [J]. Biotechnology Bulletin, 2023, 39(3): 267-277. |
| [13] | YU Shi-zhou, CAO Ling-gai, WANG Shi-ze, LIU Yong, BIAN Wen-jie, REN Xue-liang. Development Core SNP Markers for Tobacco Germplasm Genotyping [J]. Biotechnology Bulletin, 2023, 39(3): 89-100. |
| [14] | DU Qing-jie, ZHOU Lu-yao, YANG Si-zhen, ZHANG Jia-xin, CHEN Chun-lin, LI Juan-qi, LI Meng, ZHAO Shi-wen, XIAO Huai-juan, WANG Ji-qing. Overexpression of CaCP1 Enhances Salt Stress Sensibility in Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(2): 172-182. |
| [15] | WANG Ge-ge, QIU Shi-rui, ZHANG Lin-han, YANG Guo-wei, XU Xiao-yun, WANG Ai-ling, ZENG Shu-hua, LIU Ya-jie. Molecular Cytology at Meiosis in Allotriploid Nicotiana tabacum(SST) [J]. Biotechnology Bulletin, 2023, 39(2): 183-192. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||