








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (10): 93-106.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0486
Previous Articles Next Articles
LIU Jin-sheng( ), CHEN Zhen-ya(
), CHEN Zhen-ya( ), HUO Yi-xin, GUO Shu-yuan(
), HUO Yi-xin, GUO Shu-yuan( )
)
Received:2023-05-23
Online:2023-10-26
Published:2023-11-28
Contact:
CHEN Zhen-ya, GUO Shu-yuan
E-mail:ljs15536837748@163.com;chenzhenya@bit.edu.cn;guosy@bit.edu.cn
LIU Jin-sheng, CHEN Zhen-ya, HUO Yi-xin, GUO Shu-yuan. Application of FACS Technology in the Directed Evolution of Enzyme[J]. Biotechnology Bulletin, 2023, 39(10): 93-106.
| 筛选方法 Screening method | 通量 Throughput | 突变文库 Mutant library | 特点 Characteristic | 参考文献 References |
|---|---|---|---|---|
| 平板 | 105-106 | 操作简便;半定量分析;通量低;精度低 | [ | |
| 微孔板 | 1/s | 105 | 操作简便;定量分析;通量低;精度低 | [ |
| FACS | 104/s | >108 | 通量高;消耗少;精度高;单细胞定量分析;设备昂贵;不便携 | [ |
| FADS | <103/s | 107 | 通量高;消耗少;精度高;单细胞定量分析;工艺复杂度高;搭建难 | [ |
Table 1 Comparison of screening methods
| 筛选方法 Screening method | 通量 Throughput | 突变文库 Mutant library | 特点 Characteristic | 参考文献 References |
|---|---|---|---|---|
| 平板 | 105-106 | 操作简便;半定量分析;通量低;精度低 | [ | |
| 微孔板 | 1/s | 105 | 操作简便;定量分析;通量低;精度低 | [ |
| FACS | 104/s | >108 | 通量高;消耗少;精度高;单细胞定量分析;设备昂贵;不便携 | [ |
| FADS | <103/s | 107 | 通量高;消耗少;精度高;单细胞定量分析;工艺复杂度高;搭建难 | [ |
| 类别 Classification | 目标酶 Target enzyme | 结果 Result | 宿主细胞 Host cell | 参考文献 Reference |
|---|---|---|---|---|
| 细胞表面酶 | 丝氨酸蛋白酶OmpT | kcat/Km值提高60倍 | 大肠杆菌E. coli | [ |
| 分选酶A | kcat/Km值提高140倍 | 酿酒酵母S. cerevisiae | [ | |
| 半乳糖氧化酶 | Km值降低4.4倍 | 大肠杆菌E. coli | [ | |
| D-氨基酸氧化酶 | kcat值提高4.2倍 | 大肠杆菌E. coli | [ | |
| 胞内酶 | β-1,3-半乳糖基转移酶 | 底物耐受性提高300倍 | 大肠杆菌E. coli | [ |
| α-1,3-岩藻糖基转移酶 | 合成Lewis X和3'岩藻糖基乳糖的kcat/Km值分别提高6倍和14倍 | 幽门螺旋杆菌Helicobacter pylori | [ | |
| α-1,2-岩藻糖基转移酶 | 2'岩藻糖基乳糖的滴度和产量分别提高1.72倍和1.51倍 | 大肠杆菌E. coli | [ | |
| 多聚唾液酸转移酶 | 比酶活和热稳定性分别提高 1.5-2.1倍和9倍 | 奈瑟菌Neisseria | [ | |
| 葡萄糖氧化酶 | Vmax值分别提高1.3和2.3倍 | 酿酒酵母S. cerevisiae | [ | |
| ATP磷酸核糖转移酶 | L-组氨酸产量为0.1 mmol/L,野生型未生产L-组氨酸 | 谷氨酸棒状杆菌Corynebacterium glutamicum | [ | |
| 丝氨酸乙酰转移酶 | L-半胱氨酸产量提高2.36倍 | 大肠杆菌E. coli | [ | |
| 3C蛋白酶 | 蛋白水解活性提高4倍 | 大肠杆菌E. coli | [ | |
| 咖啡因去甲基化酶 | Vmax/Km值和产物选择性分别提高33和22倍 | 酿酒酵母S. cerevisiae | [ | |
| 丙酮酸羧化酶 | 赖氨酸滴度分别增加9%和19% | 谷氨酸棒状杆菌C. glutamicum | [ | |
| 莽草酸生产菌株 | 莽草酸滴度达到(3.72±0.35)mmol/L,比野生型提高2.4倍 | 谷氨酸棒状杆菌C. glutamicum | [ | |
| 丝氨酸羟甲基转移酶 | L-丝氨酸产量达到34.78 g/L,比野生型菌株提高35.9% | 谷氨酸棒状杆菌C. glutamicum | [ |
Table 2 Application of FACS in non-secretory enzymes
| 类别 Classification | 目标酶 Target enzyme | 结果 Result | 宿主细胞 Host cell | 参考文献 Reference |
|---|---|---|---|---|
| 细胞表面酶 | 丝氨酸蛋白酶OmpT | kcat/Km值提高60倍 | 大肠杆菌E. coli | [ |
| 分选酶A | kcat/Km值提高140倍 | 酿酒酵母S. cerevisiae | [ | |
| 半乳糖氧化酶 | Km值降低4.4倍 | 大肠杆菌E. coli | [ | |
| D-氨基酸氧化酶 | kcat值提高4.2倍 | 大肠杆菌E. coli | [ | |
| 胞内酶 | β-1,3-半乳糖基转移酶 | 底物耐受性提高300倍 | 大肠杆菌E. coli | [ |
| α-1,3-岩藻糖基转移酶 | 合成Lewis X和3'岩藻糖基乳糖的kcat/Km值分别提高6倍和14倍 | 幽门螺旋杆菌Helicobacter pylori | [ | |
| α-1,2-岩藻糖基转移酶 | 2'岩藻糖基乳糖的滴度和产量分别提高1.72倍和1.51倍 | 大肠杆菌E. coli | [ | |
| 多聚唾液酸转移酶 | 比酶活和热稳定性分别提高 1.5-2.1倍和9倍 | 奈瑟菌Neisseria | [ | |
| 葡萄糖氧化酶 | Vmax值分别提高1.3和2.3倍 | 酿酒酵母S. cerevisiae | [ | |
| ATP磷酸核糖转移酶 | L-组氨酸产量为0.1 mmol/L,野生型未生产L-组氨酸 | 谷氨酸棒状杆菌Corynebacterium glutamicum | [ | |
| 丝氨酸乙酰转移酶 | L-半胱氨酸产量提高2.36倍 | 大肠杆菌E. coli | [ | |
| 3C蛋白酶 | 蛋白水解活性提高4倍 | 大肠杆菌E. coli | [ | |
| 咖啡因去甲基化酶 | Vmax/Km值和产物选择性分别提高33和22倍 | 酿酒酵母S. cerevisiae | [ | |
| 丙酮酸羧化酶 | 赖氨酸滴度分别增加9%和19% | 谷氨酸棒状杆菌C. glutamicum | [ | |
| 莽草酸生产菌株 | 莽草酸滴度达到(3.72±0.35)mmol/L,比野生型提高2.4倍 | 谷氨酸棒状杆菌C. glutamicum | [ | |
| 丝氨酸羟甲基转移酶 | L-丝氨酸产量达到34.78 g/L,比野生型菌株提高35.9% | 谷氨酸棒状杆菌C. glutamicum | [ |
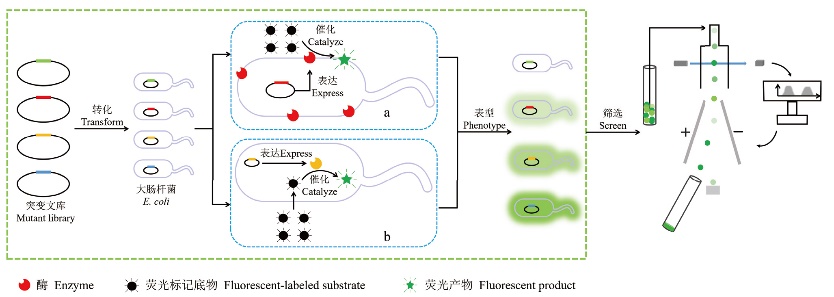
Fig. 2 FACS screening of non-secretory enzymes a: Screening of cell surface enzymes with fluorescent-labeled substrates. b: Screening of intracellular enzymes with fluorescent-labeled substrates
| 类别 Classification | 目标酶 Target enzyme | 结果 Result | 宿主细胞 Host cell | 参考文献 Reference |
|---|---|---|---|---|
| 双乳化液滴法 | 血清对氧磷酶 | kcat/Km值提高100倍 | 大肠杆菌E. coli | [ |
| 碱性磷酸酶 | 实现活性碱性磷酸酶的100万倍富集 | 大肠杆菌E. coli | [ | |
| α-L-苏糖核酸聚合酶 | 获得一个不依赖于锰的α-L-苏氨酸核糖核酸(TNA)聚合酶突变体 | 大肠杆菌E. coli | [ | |
| 几丁质酶A | 比酶活提高2倍 | 大肠杆菌E. coli | [ | |
| 人工金属酶 | 建立FACS筛选双乳化液滴中人工金属酶的方法 | 大肠杆菌E. coli | [ | |
| 水凝胶微珠法 | RNA聚合酶 | 获得一个对利福霉素有耐药性的突变体 | 大肠杆菌E. coli | [ |
| 植酸酶 | kcat值提高31% | 大肠杆菌E. coli | [ | |
| 木聚糖酶 | 比酶活提高1.3倍 | 毕赤酵母Pichia pastoris | [ | |
| 转肽酶 | kcat/Km值提高114倍 | 大肠杆菌E. coli | [ |
Table 3 Application of FACS in secretory enzymes
| 类别 Classification | 目标酶 Target enzyme | 结果 Result | 宿主细胞 Host cell | 参考文献 Reference |
|---|---|---|---|---|
| 双乳化液滴法 | 血清对氧磷酶 | kcat/Km值提高100倍 | 大肠杆菌E. coli | [ |
| 碱性磷酸酶 | 实现活性碱性磷酸酶的100万倍富集 | 大肠杆菌E. coli | [ | |
| α-L-苏糖核酸聚合酶 | 获得一个不依赖于锰的α-L-苏氨酸核糖核酸(TNA)聚合酶突变体 | 大肠杆菌E. coli | [ | |
| 几丁质酶A | 比酶活提高2倍 | 大肠杆菌E. coli | [ | |
| 人工金属酶 | 建立FACS筛选双乳化液滴中人工金属酶的方法 | 大肠杆菌E. coli | [ | |
| 水凝胶微珠法 | RNA聚合酶 | 获得一个对利福霉素有耐药性的突变体 | 大肠杆菌E. coli | [ |
| 植酸酶 | kcat值提高31% | 大肠杆菌E. coli | [ | |
| 木聚糖酶 | 比酶活提高1.3倍 | 毕赤酵母Pichia pastoris | [ | |
| 转肽酶 | kcat/Km值提高114倍 | 大肠杆菌E. coli | [ |
| 类别 Classification | 材料 Material | 特点 Characteristic | 参考文献 References |
|---|---|---|---|
| 天然水凝胶 | 纤维蛋白;胶原蛋白;海藻酸盐;透明质酸;壳聚糖;琼脂糖 | 通过物理或离子相互作用力形成凝胶;良好的生物相容性 | [ |
| 合成水凝胶 | 聚乙二醇;聚丙烯酸;聚乙烯醇 | 通过共价键形成聚合体;可再生的;易获得 |
Table 4 Types of hydrogels
| 类别 Classification | 材料 Material | 特点 Characteristic | 参考文献 References |
|---|---|---|---|
| 天然水凝胶 | 纤维蛋白;胶原蛋白;海藻酸盐;透明质酸;壳聚糖;琼脂糖 | 通过物理或离子相互作用力形成凝胶;良好的生物相容性 | [ |
| 合成水凝胶 | 聚乙二醇;聚丙烯酸;聚乙烯醇 | 通过共价键形成聚合体;可再生的;易获得 |
| [1] |
Ur Rahman A, Khan S, Khan M. Transport of trans-activator of transcription(TAT)peptide in tumour tissue model: evaluation of factors affecting the transport of TAT evidenced by flow cytometry[J]. Journal of Pharmacy and Pharmacology, 2020, 72(4): 519-530.
doi: 10.1111/jphp.13221 pmid: 31868235 |
| [2] |
Suo YZ, Gu ZQ, Wei XB. Advances of in vivo flow cytometry on cancer studies[J]. Cytometry A, 2020, 97(1): 15-23.
doi: 10.1002/cyto.a.v97.1 URL |
| [3] |
Burnie J, Tang VA, Welsh JA, et al. Flow virometry quantification of host proteins on the surface of HIV-1 pseudovirus particles[J]. Viruses, 2020, 12(11): 1296.
doi: 10.3390/v12111296 URL |
| [4] |
El-Hajjar L, Ali Ahmad F, Nasr R. A guide to flow cytometry: components, basic principles, experimental design, and cancer research applications[J]. Curr Protoc, 2023, 3(3): e721.
doi: 10.1002/cpz1.v3.3 URL |
| [5] | Domínguez LM, Fiore EJ, Mazzolini GD. Generation and characterization of human mesenchymal stem/stromal cells for cell therapy applications[J]. Methods Cell Biol, 2022, 170: 189-202. |
| [6] |
Midani FS, David LA. Tracking defined microbial communities by multicolor flow cytometry reveals tradeoffs between productivity and diversity[J]. Front Microbiol, 2023, 13: 910390.
doi: 10.3389/fmicb.2022.910390 URL |
| [7] | Malone MK, Ujas TA, Cotter KM, et al. FACS to identify immune subsets in mouse brain and spleen[M]//Methods in Molecular Biology. New York, NY: Springer US, 2023: 213-229. |
| [8] |
Becker S, Schmoldt HU, Adams TM, et al. Ultra-high-throughput screening based on cell-surface display and fluorescence-activated cell sorting for the identification of novel biocatalysts[J]. Curr Opin Biotechnol, 2004, 15(4): 323-329.
doi: 10.1016/j.copbio.2004.06.001 URL |
| [9] |
Zeymer C, Hilvert D. Directed evolution of protein catalysts[J]. Annu Rev Biochem, 2018, 87: 131-157.
doi: 10.1146/annurev-biochem-062917-012034 pmid: 29494241 |
| [10] |
Sellés Vidal L, Isalan M, Heap JT, et al. A primer to directed evolution: current methodologies and future directions[J]. RSC Chem Biol, 2023, 4(4): 271-291.
doi: 10.1039/D2CB00231K URL |
| [11] |
Wang YJ, Xue P, Cao MF, et al. Directed evolution: methodologies and applications[J]. Chem Rev, 2021, 121(20): 12384-12444.
doi: 10.1021/acs.chemrev.1c00260 pmid: 34297541 |
| [12] |
Zhu HL, Zhang HQ, Xu Y, et al. PCR past, present and future[J]. BioTechniques, 2020, 69(4): 317-325.
doi: 10.2144/btn-2020-0057 URL |
| [13] |
Chua JPS, Go MK, Osothprarop T, et al. Evolving a thermostable terminal deoxynucleotidyl transferase[J]. ACS Synth Biol, 2020, 9(7): 1725-1735.
doi: 10.1021/acssynbio.0c00078 pmid: 32497424 |
| [14] |
Hulett HR, Bonner WA, Barrett J, et al. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence[J]. Science, 1969, 166(3906): 747-749.
pmid: 4898615 |
| [15] |
Parks DR, Hardy RR, Herzenberg LA. Three-color immunofluorescence analysis of mouse B-lymphocyte subpopulations[J]. Cytometry, 1984, 5(2): 159-168.
pmid: 6370630 |
| [16] | Schweingruber C, Nijssen J, Benitez JA, et al. Single-cell mRNA-seq of in vitro-derived human neurons using smart-Seq2[M]//Transcription Factor Regulatory Networks. New York, NY: Springer US, 2022: 143-164. |
| [17] | McKinnon KM. Flow cytometry: an overview[J]. Curr Protoc Immunol, 2018, 120: 5.1.1-5.1.11. |
| [18] |
Bonilla DL, Reinin G, Chua E. Full spectrum flow cytometry as a powerful technology for cancer immunotherapy research[J]. Front Mol Biosci, 2021, 7: 612801.
doi: 10.3389/fmolb.2020.612801 URL |
| [19] |
Krasteva D, Ivanov Y, Chengolova Z, et al. Simultaneous enumeration of CD34+ and CD45+ cells using EasyCounter image cytometer[J]. Anal Biochem, 2021, 632: 114351.
doi: 10.1016/j.ab.2021.114351 URL |
| [20] |
Wang JH, Tu CG, Zhang H, et al. Loading of metal isotope-containing intercalators for mass cytometry-based high-throughput quantitation of exosome uptake at the single-cell level[J]. Biomaterials, 2020, 255: 120152.
doi: 10.1016/j.biomaterials.2020.120152 URL |
| [21] |
Li LJ, Yu SJ, Liu SS, et al. The expression and clinical significance of CD59 and FLAER in Chinese adult AML patients[J]. J Clin Lab Anal, 2022, 36(1): e24145.
doi: 10.1002/jcla.v36.1 URL |
| [22] | Lin DF, Shen LS, Luo M, et al. Circulating tumor cells: biology and clinical significance[J]. Signal Transduct Target Ther, 2021, 6(1): 404. |
| [23] | Freen-van Heeren JJ. Flow-FISH as a tool for studying bacteria, fungi and viruses[J]. BioTech(Basel), 2021, 10(4): 21. |
| [24] |
Cheng JL, Mao X, Chen CX, et al. Monitoring anti-CD19 chimeric antigen receptor T cell population by flow cytometry and its consistency with digital droplet polymerase chain reaction[J]. Cytometry A, 2023, 103(1): 16-26.
doi: 10.1002/cyto.a.v103.1 URL |
| [25] | Xiao J, Qiu M, Long MZ, et al. Long non-coding RNA XIST impedes LPS-induced AC16 cell inflammation and apoptosis through down-regulating miR-370-3p and regulating PI3K/AKT/mTOR pathways[J]. Comb Chem High Throughput Screen, 2023. DOI.org/10.2174/1386207326666230213124031. |
| [26] | 陈林, 宋丽. 流式细胞术的发展及在植物研究中的应用[J]. 生物工程学报, 2023, 39(2): 472-487. |
| Chen L, Song L. Development of flow cytometry and its application in plant research[J]. Chin J Biotechnol, 2023, 39(2): 472-487. | |
| [27] |
Shrirao AB, Fritz Z, Novik EM, et al. Microfluidic flow cytometry: the role of microfabrication methodologies, performance and functional specification[J]. Technology, 2018, 6(1): 1-23.
doi: 10.1142/S2339547818300019 pmid: 29682599 |
| [28] |
Li ZJ, Li PY, Xu J, et al. Hydrodynamic flow cytometer performance enhancement by two-dimensional acoustic focusing[J]. Biomed Microdevices, 2020, 22(2): 27.
doi: 10.1007/s10544-020-00481-9 pmid: 32222836 |
| [29] |
Robinson JP. Flow cytometry: past and future[J]. BioTechniques, 2022, 72(4): 159-169.
doi: 10.2144/btn-2022-0005 pmid: 35369735 |
| [30] |
Manohar SM, Shah P, Nair A. Flow cytometry: principles, applications and recent advances[J]. Bioanalysis, 2021, 13(3): 181-198.
doi: 10.4155/bio-2020-0267 pmid: 33543666 |
| [31] | Maby P, Corneau A, Galon J. Phenotyping of tumor infiltrating immune cells using mass-cytometry(CyTOF)[J]. Methods Enzymol, 2020, 632: 339-368. |
| [32] | Robinson JP. Spectral flow cytometry—Quo vadimus?[J]. Cytometry, 2019: cyto.a.23779. |
| [33] |
Liu HS, Tian Y, Xue CF, et al. Analysis of extracellular vesicle DNA at the single-vesicle level by nano-flow cytometry[J]. J Extracell Vesicles, 2022, 11(4): e12206.
doi: 10.1002/jev2.v11.4 URL |
| [34] |
Chaganti LK, Venkatakrishnan N, Bose K. An efficient method for FITC labelling of proteins using tandem affinity purification[J]. Biosci Rep, 2018, 38(6): BSR20181764.
doi: 10.1042/BSR20181764 URL |
| [35] |
Li JC, Pu KY. Development of organic semiconducting materials for deep-tissue optical imaging, phototherapy and photoactivation[J]. Chem Soc Rev, 2019, 48(1): 38-71.
doi: 10.1039/c8cs00001h pmid: 30387803 |
| [36] |
Molaei MJ. A review on nanostructured carbon quantum dots and their applications in biotechnology, sensors, and chemiluminescence[J]. Talanta, 2019, 196: 456-478.
doi: S0039-9140(18)31309-2 pmid: 30683392 |
| [37] |
Saini DK, Pabbi S, Shukla P. Cyanobacterial pigments: perspectives and biotechnological approaches[J]. Food Chem Toxicol, 2018, 120: 616-624.
doi: S0278-6915(18)30509-X pmid: 30077705 |
| [38] |
Wang MJ, Da YF, Tian Y. Fluorescent proteins and genetically encoded biosensors[J]. Chem Soc Rev, 2023, 52(4): 1189-1214.
doi: 10.1039/d2cs00419d pmid: 36722390 |
| [39] |
Eilenberger C, Spitz S, Bachmann BEM, et al. The usual suspects 2019: of chips, droplets, synthesis, and artificial cells[J]. Micromachines, 2019, 10(5): 285.
doi: 10.3390/mi10050285 URL |
| [40] |
Arbige MV, Shetty JK, Chotani GK. Industrial enzymology: the next chapter[J]. Trends Biotechnol, 2019, 37(12): 1355-1366.
doi: S0167-7799(19)30242-2 pmid: 31679826 |
| [41] |
Sheldon RA, Woodley JM. Role of biocatalysis in sustainable chemistry[J]. Chem Rev, 2018, 118(2): 801-838.
doi: 10.1021/acs.chemrev.7b00203 pmid: 28876904 |
| [42] |
Victorino da Silva Amatto I, Gonsales da Rosa-Garzon N, Antônio de Oliveira Simões F, et al. Enzyme engineering and its industrial applications[J]. Biotechnol Appl Biochem, 2022, 69(2): 389-409.
doi: 10.1002/bab.v69.2 URL |
| [43] | Matuła K, Rivello F, Huck WTS. Single-cell analysis using droplet microfluidics[J]. Adv Biosyst, 2020, 4(1): e1900188. |
| [44] |
Weiβ MS, Bornscheuer UT, Höhne M. Solid-phase agar plate assay for screening amine transaminases[J]. Methods Mol Biol, 2018, 1685: 283-296.
doi: 10.1007/978-1-4939-7366-8_17 pmid: 29086316 |
| [45] |
Karnaouri A, Zerva A, Christakopoulos P, et al. Screening of recombinant lignocellulolytic enzymes through rapid plate assays[J]. Methods Mol Biol, 2021, 2178: 479-503.
doi: 10.1007/978-1-0716-0775-6_30 pmid: 33128767 |
| [46] |
Jankowski N, Koschorreck K. Agar plate assay for rapid screening of aryl-alcohol oxidase mutant libraries in Pichia pastoris[J]. J Biotechnol, 2022, 346: 47-51.
doi: 10.1016/j.jbiotec.2022.01.006 pmid: 35122934 |
| [47] |
Diep P, Mahadevan R, Yakunin AF. A microplate screen to estimate metal-binding affinities of metalloproteins[J]. Anal Biochem, 2020, 609: 113836.
doi: 10.1016/j.ab.2020.113836 URL |
| [48] |
Koh DWS, Tay JH, Gan SKE. Engineering Ag43 signal peptides with bacterial display and selection[J]. Methods Protoc, 2022, 6(1): 1.
doi: 10.3390/mps6010001 URL |
| [49] |
Ma FQ, Chung MT, Yao Y, et al. Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform[J]. Nat Commun, 2018, 9(1): 1030.
doi: 10.1038/s41467-018-03492-6 pmid: 29531246 |
| [50] |
Jiang JJ, Yang GY, Ma FQ. Fluorescence coupling strategies in fluorescence-activated droplet sorting(FADS)for ultrahigh-throughput screening of enzymes, metabolites, and antibodies[J]. Biotechnol Adv, 2023, 66: 108173.
doi: 10.1016/j.biotechadv.2023.108173 URL |
| [51] |
Liu YF, Yuan HL, Ding DQ, et al. Establishment of a biosensor-based high-throughput screening platform for tryptophan overproduction[J]. ACS Synth Biol, 2021, 10(6): 1373-1383.
doi: 10.1021/acssynbio.0c00647 pmid: 34081459 |
| [52] |
Lim J, Petersen M, Bunz M, et al. Flow cytometry based-FRET: basics, novel developments and future perspectives[J]. Cell Mol Life Sci, 2022, 79(4): 217.
doi: 10.1007/s00018-022-04232-2 pmid: 35352201 |
| [53] |
Keerthana S, Sam B, George L, et al. Fluorescein based fluorescence sensors for the selective sensing of various analytes[J]. J Fluoresc, 2021, 31(5): 1251-1276.
doi: 10.1007/s10895-021-02770-9 pmid: 34255257 |
| [54] |
Olsen MJ, Stephens D, Griffiths D, et al. Function-based isolation of novel enzymes from a large library[J]. Nat Biotechnol, 2000, 18(10): 1071-1074.
pmid: 11017045 |
| [55] |
Chen I, Dorr BM, Liu DR. A general strategy for the evolution of bond-forming enzymes using yeast display[J]. Proc Natl Acad Sci USA, 2011, 108(28): 11399-11404.
doi: 10.1073/pnas.1101046108 pmid: 21697512 |
| [56] |
Feng LL, Gao L, Besirlioglu V, et al. A flow cytometry-based ultrahigh-throughput screening method for directed evolution of oxidases[J]. Angew Chem Int Ed Engl, 2023, 62(22): e202214999.
doi: 10.1002/anie.v62.22 URL |
| [57] |
Yang GY, Rich JR, Gilbert M, et al. Fluorescence activated cell sorting as a general ultra-high-throughput screening method for directed evolution of glycosyltransferases[J]. J Am Chem Soc, 2010, 132(30): 10570-10577.
doi: 10.1021/ja104167y pmid: 20662530 |
| [58] |
Tan YM, Zhang Y, Han YB, et al. Directed evolution of an α1, 3-fucosyltransferase using a single-cell ultrahigh-throughput screening method[J]. Sci Adv, 2019, 5(10): eaaw8451.
doi: 10.1126/sciadv.aaw8451 URL |
| [59] |
Shin J, Kim S, Park W, et al. Directed evolution of soluble α-1, 2-fucosyltransferase using kanamycin resistance protein as a phenotypic reporter for efficient production of 2'-fucosyllactose[J]. J Microbiol Biotechnol, 2022, 32(11): 1471-1478.
doi: 10.4014/jmb.2209.09018 URL |
| [60] |
Janesch B, Baumann L, Mark A, et al. Directed evolution of bacterial polysialyltransferases[J]. Glycobiology, 2019, 29(7): 588-598.
doi: 10.1093/glycob/cwz021 pmid: 30976781 |
| [61] |
Kovačević G, Ostafe R, Balaž AM, et al. Development of GFP-based high-throughput screening system for directed evolution of glucose oxidase[J]. J Biosci Bioeng, 2019, 127(1): 30-37.
doi: S1389-1723(18)30312-8 pmid: 30033354 |
| [62] |
Della Corte D, van Beek HL, Syberg F, et al. Engineering and application of a biosensor with focused ligand specificity[J]. Nat Commun, 2020, 11(1): 4851.
doi: 10.1038/s41467-020-18400-0 pmid: 32978386 |
| [63] |
Gao JS, Du MH, Zhao JH, et al. Design of a genetically encoded biosensor to establish a high-throughput screening platform for L-cysteine overproduction[J]. Metab Eng, 2022, 73: 144-157.
doi: 10.1016/j.ymben.2022.07.007 pmid: 35921946 |
| [64] |
Meister SW, Hendrikse NM, Löfblom J. Directed evolution of the 3C protease from coxsackievirus using a novel fluorescence-assisted intracellular method[J]. Biol Chem, 2019, 400(3): 405-415.
doi: 10.1515/hsz-2018-0362 pmid: 30521472 |
| [65] |
Michener JK, Smolke CD. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch[J]. Metab Eng, 2012, 14(4): 306-316.
doi: 10.1016/j.ymben.2012.04.004 pmid: 22554528 |
| [66] |
Kortmann M, Mack C, Baumgart M, et al. Pyruvate carboxylase variants enabling improved lysine production from glucose identified by biosensor-based high-throughput fluorescence-activated cell sorting screening[J]. ACS Synth Biol, 2019, 8(2): 274-281.
doi: 10.1021/acssynbio.8b00510 pmid: 30707564 |
| [67] |
Liu C, Zhang B, Liu YM, et al. New intracellular shikimic acid biosensor for monitoring shikimate synthesis in Corynebacterium glutamicum[J]. ACS Synth Biol, 2018, 7(2): 591-601.
doi: 10.1021/acssynbio.7b00339 URL |
| [68] |
Zhang X, Zhang XM, Xu GQ, et al. Integration of ARTP mutagenesis with biosensor-mediated high-throughput screening to improve L-serine yield in Corynebacterium glutamicum[J]. Appl Microbiol Biotechnol, 2018, 102(14): 5939-5951.
doi: 10.1007/s00253-018-9025-2 pmid: 29725721 |
| [69] |
Yang GY, Withers SG. Ultrahigh-throughput FACS-based screening for directed enzyme evolution[J]. Chembiochem, 2009, 10(17): 2704-2715.
doi: 10.1002/cbic.200900384 pmid: 19780076 |
| [70] |
Zhou GJ, Zhang FZ. Applications and tuning strategies for transcription factor-based metabolite biosensors[J]. Biosensors, 2023, 13(4): 428.
doi: 10.3390/bios13040428 URL |
| [71] |
Mohamed M G A, Ambhorkar P, Samanipour R, et al. Microfluidics-based fabrication of cell-laden microgels[J]. Biomicrofluidics, 2020, 14(2): 021501.
doi: 10.1063/1.5134060 URL |
| [72] |
Brower KK, Khariton M, Suzuki PH, et al. Double emulsion picoreactors for high-throughput single-cell encapsulation and phenotyping via FACS[J]. Anal Chem, 2020, 92(19): 13262-13270.
doi: 10.1021/acs.analchem.0c02499 URL |
| [73] | Schaerli Y. Bacterial microcolonies in gel beads for high-throughput screening[J]. Bio-protocol, 2018, 8(13): e2911. |
| [74] |
Aharoni A, Amitai G, Bernath K, et al. High-throughput screening of enzyme libraries: thiolactonases evolved by fluorescence-activated sorting of single cells in emulsion compartments[J]. Chem Biol, 2005, 12(12): 1281-1289.
doi: 10.1016/j.chembiol.2005.09.012 URL |
| [75] |
Zinchenko A, Devenish SRA, Kintses B, et al. One in a million: flow cytometric sorting of single cell-lysate assays in monodisperse picolitre double emulsion droplets for directed evolution[J]. Anal Chem, 2014, 86(5): 2526-2533.
doi: 10.1021/ac403585p pmid: 24517505 |
| [76] |
Larsen AC, Dunn MR, Hatch A, et al. A general strategy for expanding polymerase function by droplet microfluidics[J]. Nat Commun, 2016, 7: 11235.
doi: 10.1038/ncomms11235 pmid: 27044725 |
| [77] |
Menghiu G, Ostafe V, Prodanović R, et al. A high-throughput screening system based on fluorescence-activated cell sorting for the directed evolution of chitinase A[J]. Int J Mol Sci, 2021, 22(6): 3041.
doi: 10.3390/ijms22063041 URL |
| [78] |
Vallapurackal J, Stucki A, Liang AD, et al. Ultrahigh-throughput screening of an artificial metalloenzyme using double emulsions[J]. Angew Chem Int Ed Engl, 2022, 61(48): e202207328.
doi: 10.1002/anie.v61.48 URL |
| [79] |
Eun YJ, Utada AS, Copeland MF, et al. Encapsulating bacteria in agarose microparticles using microfluidics for high-throughput cell analysis and isolation[J]. ACS Chem Biol, 2011, 6(3): 260-266.
doi: 10.1021/cb100336p URL |
| [80] |
Pitzler C, Wirtz G, Vojcic L, et al. A fluorescent hydrogel-based flow cytometry high-throughput screening platform for hydrolytic enzymes[J]. Chem Biol, 2014, 21(12): 1733-1742.
doi: 10.1016/j.chembiol.2014.10.018 URL |
| [81] |
Ma CX, Tan ZL, Lin Y, et al. Gel microdroplet-based high-throughput screening for directed evolution of xylanase-producing Pichia pastoris[J]. J Biosci Bioeng, 2019, 128(6): 662-668.
doi: 10.1016/j.jbiosc.2019.05.008 URL |
| [82] |
Gianella P, Snapp EL, Levy M. An in vitro compartmentalization-based method for the selection of bond-forming enzymes from large libraries[J]. Biotechnol Bioeng, 2016, 113(8): 1647-1657.
doi: 10.1002/bit.25939 pmid: 26806853 |
| [83] |
Bernath K, Hai MT, Mastrobattista E, et al. In vitro compartmentalization by double emulsions: sorting and gene enrichment by fluorescence activated cell sorting[J]. Anal Biochem, 2004, 325(1): 151-157.
pmid: 14715296 |
| [84] |
Watanabe T, Motohiro I, Ono T. Microfluidic formation of hydrogel microcapsules with a single aqueous core by spontaneous cross-linking in aqueous two-phase system droplets[J]. Langmuir, 2019, 35(6): 2358-2367.
doi: 10.1021/acs.langmuir.8b04169 pmid: 30626189 |
| [85] |
Lai EP, Bao BY, Zhu YF, et al. Transglutaminase-catalyzed bottom-up synthesis of polymer hydrogel[J]. Front Bioeng Biotechnol, 2022, 10: 824747.
doi: 10.3389/fbioe.2022.824747 URL |
| [86] |
Zhan J, Sun HY, Xie MM, et al. Hyperbranched polyamidoamine-chitosan polyelectrolyte gels crosslinking by polyacrylic acid and alginate for removal of anionic dyes[J]. Int J Biol Macromol, 2022, 222(Pt B): 3024-3033.
doi: 10.1016/j.ijbiomac.2022.10.077 URL |
| [87] |
Li M, Liu HR, Zhuang SY, et al. Droplet flow cytometry for single-cell analysis[J]. RSC Adv, 2021, 11(34): 20944-20960.
doi: 10.1039/d1ra02636d pmid: 35479393 |
| [88] |
Bilal M, Cui JD, Iqbal HMN. Tailoring enzyme microenvironment: state-of-the-art strategy to fulfill the quest for efficient bio-catalysis[J]. Int J Biol Macromol, 2019, 130: 186-196.
doi: S0141-8130(18)37157-5 pmid: 30817963 |
| [89] |
Markel U, Essani KD, Besirlioglu V, et al. Advances in ultrahigh-throughput screening for directed enzyme evolution[J]. Chem Soc Rev, 2020, 49(1): 233-262.
doi: 10.1039/c8cs00981c pmid: 31815263 |
| [90] |
Medcalf EJ, Gantz M, Kaminski TS, et al. Ultra-high-throughput absorbance-activated droplet sorting for enzyme screening at kilohertz frequencies[J]. Anal Chem, 2023, 95(10): 4597-4604.
doi: 10.1021/acs.analchem.2c04144 URL |
| [91] | 康里奇, 谈攀, 洪亮. 人工智能时代下的酶工程[J]. 合成生物学, 2023. https://kns.cnki.net/kcms/detail/10.1687.q.20230407.1430.002.html. |
| Kang LQ, Tan P, Hong L. Enzyme engineering in the age of artificial intelligence[J]. Synthetic Biology Journal, 2023. https://kns.cnki.net/kcms/detail/10.1687.q.20230407.1430.002.html. | |
| [92] | 曲玉辰, 陆路, 姜世勃. 利用I-Mutant2.0辅助设计与优化中东呼吸综合征冠状病毒融合抑制多肽[J]. 微生物与感染, 2019, 14(2): 72-81. |
| Qu YC, Lu L, Jiang SB. Using I-Mutant2.0 to assist the design and optimization of MERS-CoV fusion inhibitory peptides[J]. J Microbes Infect, 2019, 14(2): 72-81. | |
| [93] |
Yang Y, Ding XS, Zhu GC, et al. ProTstab - predictor for cellular protein stability[J]. BMC Genomics, 2019, 20(1): 804.
doi: 10.1186/s12864-019-6138-7 pmid: 31684883 |
| [94] | Meier J, Rao R, Verkuil R, et al. Language models enable zero-shot prediction of the effects of mutations on protein function[J]. Advances in Neural Information Processing Systems, 2021, 34: 29287-23303. |
| [1] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [2] | WANG Ya-li, WANG Na, CHENG Hong-mei. Comparison of Methods for Rapid Determination of Cotton Ploidy by Flow Cytometry [J]. Biotechnology Bulletin, 2022, 38(12): 144-148. |
| [3] | ZHANG Xue, TAN Yu-meng, JIANG Hai-xia, YANG Guang-yu. Directed Evolution of α-1,2-fucosyltransferase by a Single-cell Ultra-high-throughput Screening Method [J]. Biotechnology Bulletin, 2022, 38(1): 289-298. |
| [4] | ZHENG Ying-zhuan, LÜ Yan, YANG Dong-xu, LI Guo-wei, WANG Hong-yang, LI Can-hui. Study on the Identification of Potato Ploidy Using Flow Cytometry Based on Liquid Nitrogen Grinding Method [J]. Biotechnology Bulletin, 2021, 37(1): 282-288. |
| [5] | MI Liang-bo, ZENG Wei-zhu, HUANG Ke-xue, WANG De-ming, DU Guo-cheng, ZHOU Jing-wen, CHEN Jian. High-throughput Screening High-yield Bacitracin Strain from Bacillus licheniformis DW2 [J]. Biotechnology Bulletin, 2020, 36(7): 90-96. |
| [6] | CHEN Lin, PAN Zhen-zhi, DAI Yi, SONG Li. Screening and Application of the Nuclear Dissociation Solutions of Soybeans Suitable for Flow Cytometry Analysis [J]. Biotechnology Bulletin, 2020, 36(11): 230-237. |
| [7] | WANG Ya-nan, WEN Hai-ruo, WANG Xue. Establishment and Preliminary Exploration of in vitro Pig-a Gene Mutation Assay Based on L5178Y Cells [J]. Biotechnology Bulletin, 2020, 36(1): 220-228. |
| [8] | LI Yan-wei, SONG Xing-hui, WANG Jia-jia, LIU Li, HUANG Ying-ying, GUO Chun. Establishment of the Real-time and Label-free Screening System for Tumor Cell Apoptosis [J]. Biotechnology Bulletin, 2019, 35(10): 220-226. |
| [9] | HE Shuo-kang, LUO Ze-wei. Development and Phenotypic Analysis of Tetraploid Arabidopsis thaliana with QUARTET Mutation [J]. Biotechnology Bulletin, 2018, 34(7): 119-125. |
| [10] | LU Jia, DENG Qiu-ping, REN Wen-hua. Mechanism of Antimicrobial Peptide Scolopin 2-NH2 Isolated from Scolopendra subspinipes mutilans [J]. Biotechnology Bulletin, 2018, 34(11): 179-190. |
| [11] | SUN Wen, ZHENG Feng. Molecular Cloning of Gene for Enolase in Highly Virulent Strains from Streptococcus suis serotype 2 and Its Protein Biological Function [J]. Biotechnology Bulletin, 2017, 33(4): 222-230. |
| [12] | MA Fu-qiang, YANG Guang-yu. Ultra-high-throughput Screening System Based on Droplet Microfluidics and Its Applications in Synthetic Biology [J]. Biotechnology Bulletin, 2017, 33(1): 83-92. |
| [13] | ZHANG Jing1, 2, SUN Rui-qiu2, TANG Yan-ting2, LIU Xiang2. Establishment and Application of a High-throughput Drug Screening Assay Targeting Macrophage Migration Inhibitory Factor [J]. Biotechnology Bulletin, 2016, 32(9): 253-259. |
| [14] | Han Yawei, Wang Xihua, Chen Liping, Shi Guiqin, Sun Liping, Zhou Wenshan. Toxic Effects of NNK on NCTC 1469 Cells [J]. Biotechnology Bulletin, 2015, 31(9): 218-223. |
| [15] | Fang Yueqin, Guo Juanning, Lu Haojie, Zhu Guoqiang,. Construction of a Multipurpose M13KE Phage Display System [J]. Biotechnology Bulletin, 2014, 0(2): 143-147. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||