








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (10): 107-114.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0479
Previous Articles Next Articles
WAN Qi-wu1,2,4( ), BAO Xu-dong1,2, DING Ke1,3, MOU Hua-ming4(
), BAO Xu-dong1,2, DING Ke1,3, MOU Hua-ming4( ), LUO Yang1(
), LUO Yang1( )
)
Received:2023-05-18
Online:2023-10-26
Published:2023-11-28
Contact:
MOU Hua-ming, LUO Yang
E-mail:qiwuwan@163.com;luoy@cqu.edu.cn;mouhm2002@aliyun.com
WAN Qi-wu, BAO Xu-dong, DING Ke, MOU Hua-ming, LUO Yang. Research Progress in Microfluidic Technology in the Detection of Pathogenic Microorganisms[J]. Biotechnology Bulletin, 2023, 39(10): 107-114.
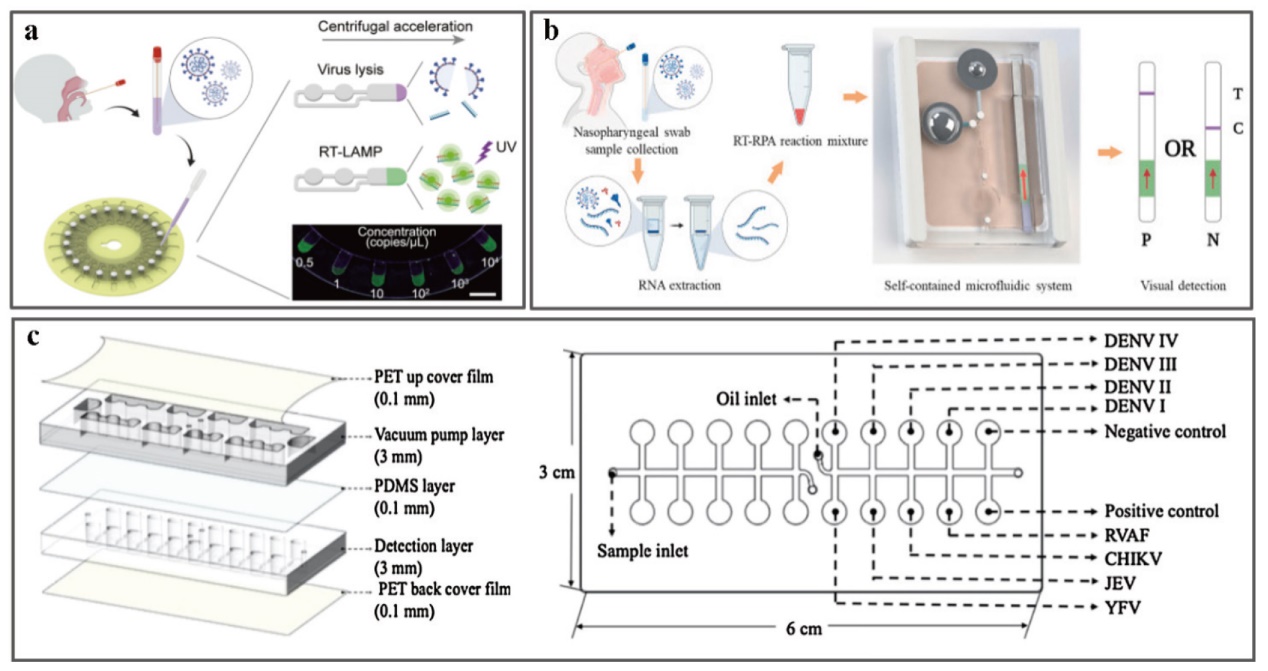
Fig. 2 Microfluidic system for virus detection a: Fully automated centrifugal microfluidic system for sample response viral nucleic acid detection[11]; b: a microfluidic system based on CRISPR diagnosis without instruments for SARS-CoV-2[12]; c: a self-powered fast-loading microfluidic chip using RT-LAMP to detect carrier viruses[13]

Fig. 3 Microfluidic system for bacterial detection a: Microfluidic platform for multiple detection of E. coli and Pseudomonas aeruginosa in blood[18]. b: Microfluidic chip for detecting drug resistance at single cell level[20]. c: Salmonella typhimurium microfluidic biosensor based on magnetic separation, enzyme catalysis and electrochemical impedance analysis[22]. d: One-pot digital microfluidic system using LNA/DNA molecular beacons for bacterial detection and absolute quantification[23]
| [1] |
Zhang DX, Bi HY, Liu BH, et al. Detection of pathogenic microorganisms by microfluidics based analytical methods[J]. Anal Chem, 2018, 90(9): 5512-5520.
doi: 10.1021/acs.analchem.8b00399 pmid: 29595252 |
| [2] |
Trinh TND, Lee NY. Advances in nucleic acid amplification-based microfluidic devices for clinical microbial detection[J]. Chemosensors, 2022, 10(4): 123.
doi: 10.3390/chemosensors10040123 URL |
| [3] |
Fernandes AC, Semenova D, Panjan P, et al. Multi-function microfluidic platform for sensor integration[J]. N Biotechnol, 2018, 47: 8-17.
doi: 10.1016/j.nbt.2018.03.001 URL |
| [4] |
Ahmed I, Akram Z, Bule M, et al. Advancements and potential applications of microfluidic approaches—a review[J]. Chemosensors, 2018, 6(4): 46.
doi: 10.3390/chemosensors6040046 URL |
| [5] |
Ait M L, Mozokhina A, Tokarev A, et al. Virus replication and competition in a cell culture: application to the SARS-CoV-2 variants[J]. Appl Math Lett, 2022, 133: 108217.
doi: 10.1016/j.aml.2022.108217 URL |
| [6] |
Li X, Zhao XY, Yang WH, et al. Stretch-driven microfluidic chip for nucleic acid detection[J]. Biotechnol Bioeng, 2021, 118(9): 3559-3568.
doi: 10.1002/bit.v118.9 URL |
| [7] |
Su WD, Qiu JJ, Mei Y, et al. A microfluidic cell chip for virus isolation via rapid screening for permissive cells[J]. Virol Sin, 2022, 37(4): 547-557.
doi: 10.1016/j.virs.2022.04.011 pmid: 35504535 |
| [8] |
Saraf N, Villegas M, Willenberg BJ, et al. Multiplex viral detection platform based on a aptamers-integrated microfluidic channel[J]. ACS Omega, 2019, 4(1): 2234-2240.
doi: 10.1021/acsomega.8b03277 pmid: 30729227 |
| [9] | Guan XJ, Wu F, Mao M, et al. A microfluidic platform for the ultrasensitive detection of human enterovirus 71[J]. Sens Actuat Rep, 2021, 3: 100046. |
| [10] |
Eivazzadeh-Keihan R, Pashazadeh-Panahi P, Mahmoudi T, et al. Dengue virus: a review on advances in detection and trends - from conventional methods to novel biosensors[J]. Mikrochim Acta, 2019, 186(6): 329.
doi: 10.1007/s00604-019-3420-y pmid: 31055654 |
| [11] |
Tian F, Liu C, Deng JQ, et al. A fully automated centrifugal microfluidic system for sample-to-answer viral nucleic acid testing[J]. Sci China Chem, 2020, 63(10): 1498-1506.
doi: 10.1007/s11426-020-9800-6 |
| [12] |
Li ZY, Ding X, Yin K, et al. Instrument-free, CRISPR-based diagnostics of SARS-CoV-2 using self-contained microfluidic system[J]. Biosens Bioelectron, 2022, 199: 113865.
doi: 10.1016/j.bios.2021.113865 URL |
| [13] |
Yao YH, Zhao N, Jing WW, et al. A self-powered rapid loading microfluidic chip for vector-borne viruses detection using RT-LAMP[J]. Sens Actuat B, 2021, 333: 129521.
doi: 10.1016/j.snb.2021.129521 URL |
| [14] |
Kim S, Akarapipad P, Nguyen BT, et al. Direct capture and smartphone quantification of airborne SARS-CoV-2 on a paper microfluidic chip[J]. Biosens Bioelectron, 2022, 200: 113912.
doi: 10.1016/j.bios.2021.113912 URL |
| [15] |
Zhang Y, Hu XZ, Wang QJ. Review of microchip analytical methods for the determination of pathogenic Escherichia coli[J]. Talanta, 2021, 232: 122410.
doi: 10.1016/j.talanta.2021.122410 URL |
| [16] |
Boehm DA, Gottlieb PA, Hua SZ. On-chip microfluidic biosensor for bacterial detection and identification[J]. Sens Actuat B, 2007, 126(2): 508-514.
doi: 10.1016/j.snb.2007.03.043 URL |
| [17] |
Mairhofer J, Roppert K, Ertl P. Microfluidic systems for pathogen sensing: a review[J]. Sensors, 2009, 9(6): 4804-4823.
doi: 10.3390/s90604804 pmid: 22408555 |
| [18] |
Costa SP, Caneira CRF, Chu V, et al. A microfluidic platform combined with bacteriophage receptor binding proteins for multiplex detection of Escherichia coli and Pseudomonas aeruginosa in blood[J]. Sens Actuat B, 2023, 376: 132917.
doi: 10.1016/j.snb.2022.132917 URL |
| [19] |
Kim S, Romero-Lozano A, Hwang DS, et al. A guanidinium-rich polymer as a new universal bioreceptor for multiplex detection of bacteria from environmental samples[J]. J Hazard Mater, 2021, 413: 125338.
doi: 10.1016/j.jhazmat.2021.125338 URL |
| [20] |
Song KN, Yu ZQ, Zu XY, et al. Microfluidic chip for detection of drug resistance at the single-cell level[J]. Micromachines, 2022, 14(1): 46.
doi: 10.3390/mi14010046 URL |
| [21] |
Sun DL, Fan TT, Liu F, et al. A microfluidic chemiluminescence biosensor based on multiple signal amplification for rapid and sensitive detection of E.coli O157: H7[J]. Biosens Bioelectron, 2022, 212: 114390.
doi: 10.1016/j.bios.2022.114390 URL |
| [22] |
Liu YJ, Jiang D, Wang SY, et al. A microfluidic biosensor for rapid detection of Salmonella typhimurium based on magnetic separation, enzymatic catalysis and electrochemical impedance analysis[J]. Chin Chem Lett, 2022, 33(6): 3156-3160.
doi: 10.1016/j.cclet.2021.10.064 URL |
| [23] |
Kao YT, Calabrese S, Borst N, et al. Microfluidic one-pot digital droplet FISH using LNA/DNA molecular beacons for bacteria detection and absolute quantification[J]. Biosensors, 2022, 12(4): 237.
doi: 10.3390/bios12040237 URL |
| [24] |
Cao DJ, Wu S, Xi CL, et al. Preparation of long single-strand DNA concatemers for high-level fluorescence in situ hybridization[J]. Commun Biol, 2021, 4(1): 1224.
doi: 10.1038/s42003-021-02762-2 |
| [25] |
Zhou WT, Le J, Chen Y, et al. Recent advances in microfluidic devices for bacteria and fungus research[J]. Trac Trends Anal Chem, 2019, 112: 175-195.
doi: 10.1016/j.trac.2018.12.024 URL |
| [26] |
Soares RRG, Santos DR, Pinto IF, et al. Point-of-use ultrafast single-step detection of food contaminants: a novel microfluidic fluorescence-based immunoassay with integrated photodetection[J]. Procedia Eng, 2016, 168: 329-332.
doi: 10.1016/j.proeng.2016.11.208 URL |
| [27] |
Schell WA, Benton JL, Smith PB, et al. Evaluation of a digital microfluidic real-time PCR platform to detect DNA of Candida albicans in blood[J]. Eur J Clin Microbiol Infect Dis, 2012, 31(9): 2237-2245.
doi: 10.1007/s10096-012-1561-6 URL |
| [28] |
Wulff-Burchfield E, Schell WA, Eckhardt AE, et al. Microfluidic platform versus conventional real-time polymerase chain reaction for the detection of Mycoplasma pneumoniae in respiratory specimens[J]. Diagn Microbiol Infect Dis, 2010, 67(1): 22-29.
doi: 10.1016/j.diagmicrobio.2009.12.020 URL |
| [29] |
Wang AY, Wu ZH, Huang YH, et al. A 3D-printed microfluidic device for qPCR detection of macrolide-resistant mutations of Mycoplasma pneumoniae[J]. Biosensors, 2021, 11(11): 427.
doi: 10.3390/bios11110427 URL |
| [30] |
Dean D, Turingan RS, Thomann HU, et al. A multiplexed microfluidic PCR assay for sensitive and specific point-of-care detection of Chlamydia trachomatis[J]. PLoS One, 2012, 7(12): e51685.
doi: 10.1371/journal.pone.0051685 URL |
| [31] |
Ye X, Li Y, Wang LJ, et al. All-in-one microfluidic nucleic acid diagnosis system for multiplex detection of sexually transmitted pathogens directly from genitourinary secretions[J]. Talanta, 2021, 221: 121462.
doi: 10.1016/j.talanta.2020.121462 URL |
| [32] |
Li L, Miao BG, Li Z, et al. Sample-to-answer hepatitis B virus DNA detection from whole blood on a centrifugal microfluidic platform with double rotation axes[J]. ACS Sens, 2019, 4(10): 2738-2745.
doi: 10.1021/acssensors.9b01270 URL |
| [1] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [2] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| [3] | SHI Jia, ZHU Xiu-mei, XUE Meng-yu, YU Chao, WEI Yi-ming, YANG Feng-huan, CHEN Hua-min. Optimization and Application of the Chromatin Immunoprecipitation Based on Rice Protoplast [J]. Biotechnology Bulletin, 2022, 38(7): 62-69. |
| [4] | MA Ya-nan, LU Xu, WEI Yun-chun, LI Kang, WEI Ruo-nan, LI Sheng, MA Shao-ying. Identification and Tissue Specific Expression Analysis of AKR Gene Family in Grape [J]. Biotechnology Bulletin, 2021, 37(8): 141-151. |
| [5] | XU Hong-yun, ZHANG En-hui, Yu Cun. Tamarix hispida Transcription Factor ThWRKY4 Binds to ARR1AT Motif to Regulate Gene Expression [J]. Biotechnology Bulletin, 2021, 37(3): 18-26. |
| [6] | LI Ling, YANG Li-xia, GUO Mei. Function of Transcription Factor CNR in the Ripening Process of Tomato Fruit [J]. Biotechnology Bulletin, 2021, 37(2): 51-62. |
| [7] | FAN Yu-chen, LU Yao, LIU Xiang-nan, ZHAO Bo. Construction of Mutants Swapping Ubiquitin E3 Ligase CHIP and E4B U-box Domain and Verification of Ubiquitination Activity [J]. Biotechnology Bulletin, 2021, 37(12): 191-197. |
| [8] | ZHAO Rui-xiao, HAN Fei, JIANG Ming-feng, ZENG Lian, HAN Jia-yu, HUANG Ying, ZHOU Hang. Application of PCR-membrane Chip Technology in the Identification of Yak and Yak-cattle Meat [J]. Biotechnology Bulletin, 2020, 36(1): 202-208. |
| [9] | LIU Na, DU Pan-pan, YANG Yang, LI Xiao-mao. Research Progress on Exosomes Isolation Methods Based on Microfluidics Technology [J]. Biotechnology Bulletin, 2019, 35(1): 207-213. |
| [10] | WANG Shuo, DING Lan, LIU Jian-xiang, HAN Jia-jia. PIF4-Regulated Thermo-responsive Genes in Arabidopsis [J]. Biotechnology Bulletin, 2018, 34(7): 57-65. |
| [11] | MA Fu-qiang, YANG Guang-yu. Ultra-high-throughput Screening System Based on Droplet Microfluidics and Its Applications in Synthetic Biology [J]. Biotechnology Bulletin, 2017, 33(1): 83-92. |
| [12] | WANG Wei-xuan, SUN Jing-yi, LIU Wei-na , HOU Ling-yu, YU Wang-ning, HE Xiang-wei, XIE Xiang-ming. The Application Progress of Microfluidic Chips in Studying Plant Cells [J]. Biotechnology Bulletin, 2016, 32(6): 19-29. |
| [13] | ZHAI Xu-zhao, WANG Guang-bin, ZHAO Liang-tao, ZENG Hai-juan, LI Jian-wu, DING Cheng-chao, SONG Chun-mei, LIU Qing. An Overview of High Throughput Biological Screening Methods and Its Application [J]. Biotechnology Bulletin, 2016, 32(6): 38-46. |
| [14] | TAO Hong,ZHANG Shao-ping,ZHANG Lin-na,ZHANG Man,HU Qi-kuan,. Screening of Stem Genes Regulated by H3K9me2 in Tumor Stem Cells [J]. Biotechnology Bulletin, 2016, 32(12): 195-202. |
| [15] | Lü Xuefeng, Abliz Ablimit Pa Naer, Tao Weidong, Xing Weiting, Cai Fula, Zheng Wenxin. Identification of Cashmere and Sheep down by Microarray Technology [J]. Biotechnology Bulletin, 2014, 0(1): 196-200. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||