








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (10): 80-92.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0271
Previous Articles Next Articles
LI Ren-han1,2( ), ZHANG Le-le1,2, LIU Chun-li1,2,3, LIU Xiu-xia1,2,3, BAI Zhong-hu1,2,3, YANG Yan-kun1,2,3(
), ZHANG Le-le1,2, LIU Chun-li1,2,3, LIU Xiu-xia1,2,3, BAI Zhong-hu1,2,3, YANG Yan-kun1,2,3( ), LI Ye1,2,3(
), LI Ye1,2,3( )
)
Received:2023-03-23
Online:2023-10-26
Published:2023-11-28
Contact:
YANG Yan-kun, LI Ye
E-mail:6200201078@stu.jiangnan.edu.cn;yangyankun@jiangnan.edu.cn;yeli0622@jiangnan.edu.cn
LI Ren-han, ZHANG Le-le, LIU Chun-li, LIU Xiu-xia, BAI Zhong-hu, YANG Yan-kun, LI Ye. Development of an L-tryptophan Biosensor Based on the Violacein Biosynthesis Pathway[J]. Biotechnology Bulletin, 2023, 39(10): 80-92.
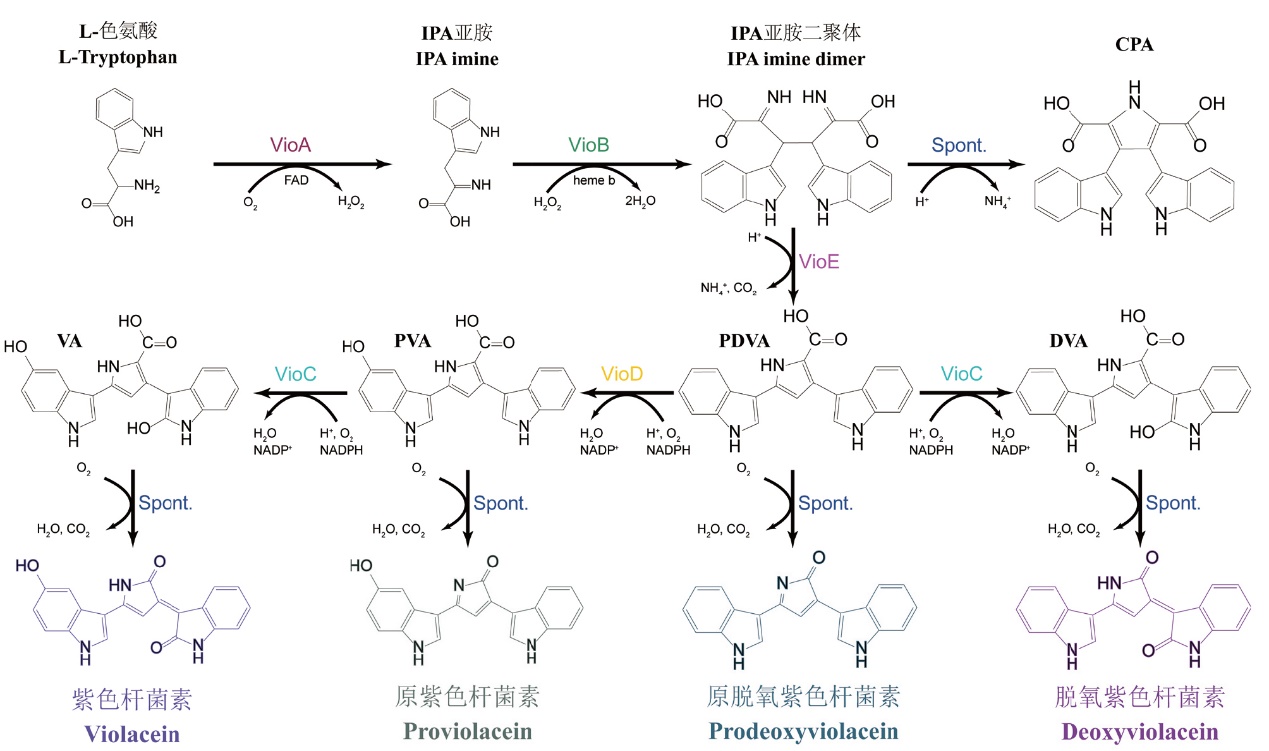
Fig. 1 Violacein biosynthetic pathway IPA: Indole-3-pyruvic acid; CPA: chromopyrrolic acid; VA: violaceinic acid; PVA: protoviolaceinic acid; PDVA: protoviolaceinic acid; DVA: deoxyviolaceinic acid; Spont: spontaneous reaction
| 质粒Plasmid | 描述Description | 来源Source |
|---|---|---|
| pVio | 在pRSFDuet载体的多克隆位点2内插入vioABCDE基因簇,卡那霉素抗性 pRSFDuet with vioABCDE gene cluster in MCS-2, KanR | [ |
| pBL_401 | 删除了pVio中的vioD基因 pVio deleted vioD | 本研究构建This study |
| pBL_403-407, 421-423 | 更换pBL_401中的RBSb区域 pBL_401 changed RBSb region | 本研究构建This study |
| pBL_415 | 将pBL_401的Duganella sp. B2 vioA基因从多克隆位点2移到多克隆位点1,在多克隆位点前添加一个T7终止子 Duganella sp. B2 vioA of pBL_401 was moved to MCS-1 from MCS-2, and a T7 terminator was added before MCS-2 | 本研究构建This study |
| pBL_416 | 将pBL_415的Duganella sp. B2 vioA替换为Janthinobacteriun violaceinigrum vioA Duganella sp. B2 vioA of pBL_415 was replaced by Janthinobacteriun violaceinigrum vioA | 本研究构建This study |
| pBL_417 | 将pBL_415的Duganella sp. B2 vioA替换为Pseudoduganella violaceinigra vioA Duganella sp. B2 vioA of pBL_415 was replaced by Pseudoduganella violaceinigra vioA | 本研究构建This study |
| pBL_418 | 将pBL_415的Duganella sp. B2 vioA替换为Myxococcus xanthus vioA Duganella sp. B2 vioA of pBL_415 was replaced by Myxococcus xanthus vioA | 本研究构建This study |
| pBL_419 | 将pBL_415的Duganella sp. B2 vioA替换为Chromobacterium violaceinum vioA Duganella sp. B2 vioA of pBL_415 was replaced by Chromobacterium violaceinum vioA | 本研究构建This study |
| pBL_420 | 将pBL_415的Duganella sp. B2 vioA替换为Duganella sp. HH105 vioA Duganella sp. B2 vioA of pBL_415 was replaced by Duganella sp. HH105 vioA | 本研究构建This study |
| pBL_425 | 更换pBL_415中的RBSa区域 pBL_415 changed RBSa region | 本研究构建This study |
| pBL_429 | 更换pBL_416中的RBSa区域 pBL_416 changed RBSa region | 本研究构建This study |
| pBL_437-439 | 更换pBL_419中的RBSa区域 pBL_419 changed RBSa region | 本研究构建This study |
| pED_RBS1-RBS4 | 携带trpEfbrD和不同RBS的pACYCDuet,氯霉素抗性 pACYCDuet with trpEfbrD and different RBSs, CmR | [ |
Table 1 Plasmids in this study
| 质粒Plasmid | 描述Description | 来源Source |
|---|---|---|
| pVio | 在pRSFDuet载体的多克隆位点2内插入vioABCDE基因簇,卡那霉素抗性 pRSFDuet with vioABCDE gene cluster in MCS-2, KanR | [ |
| pBL_401 | 删除了pVio中的vioD基因 pVio deleted vioD | 本研究构建This study |
| pBL_403-407, 421-423 | 更换pBL_401中的RBSb区域 pBL_401 changed RBSb region | 本研究构建This study |
| pBL_415 | 将pBL_401的Duganella sp. B2 vioA基因从多克隆位点2移到多克隆位点1,在多克隆位点前添加一个T7终止子 Duganella sp. B2 vioA of pBL_401 was moved to MCS-1 from MCS-2, and a T7 terminator was added before MCS-2 | 本研究构建This study |
| pBL_416 | 将pBL_415的Duganella sp. B2 vioA替换为Janthinobacteriun violaceinigrum vioA Duganella sp. B2 vioA of pBL_415 was replaced by Janthinobacteriun violaceinigrum vioA | 本研究构建This study |
| pBL_417 | 将pBL_415的Duganella sp. B2 vioA替换为Pseudoduganella violaceinigra vioA Duganella sp. B2 vioA of pBL_415 was replaced by Pseudoduganella violaceinigra vioA | 本研究构建This study |
| pBL_418 | 将pBL_415的Duganella sp. B2 vioA替换为Myxococcus xanthus vioA Duganella sp. B2 vioA of pBL_415 was replaced by Myxococcus xanthus vioA | 本研究构建This study |
| pBL_419 | 将pBL_415的Duganella sp. B2 vioA替换为Chromobacterium violaceinum vioA Duganella sp. B2 vioA of pBL_415 was replaced by Chromobacterium violaceinum vioA | 本研究构建This study |
| pBL_420 | 将pBL_415的Duganella sp. B2 vioA替换为Duganella sp. HH105 vioA Duganella sp. B2 vioA of pBL_415 was replaced by Duganella sp. HH105 vioA | 本研究构建This study |
| pBL_425 | 更换pBL_415中的RBSa区域 pBL_415 changed RBSa region | 本研究构建This study |
| pBL_429 | 更换pBL_416中的RBSa区域 pBL_416 changed RBSa region | 本研究构建This study |
| pBL_437-439 | 更换pBL_419中的RBSa区域 pBL_419 changed RBSa region | 本研究构建This study |
| pED_RBS1-RBS4 | 携带trpEfbrD和不同RBS的pACYCDuet,氯霉素抗性 pACYCDuet with trpEfbrD and different RBSs, CmR | [ |
| 菌株Strain | 描述Description | 来源 Source |
|---|---|---|
| E. coli JM109 | 用于构建质粒Construct plasmids | 实验室保存Lab stored |
| E. coli BL21(DE3) | 用于生物传感器宿主菌The host of the biosensor | 实验室保存Lab stored |
| E. coli B8 | 敲除trpR tnaA pheA的E. coli BL21(DE3)E. coli BL21(DE3)ΔtrpR ΔtnaA ΔpheA | [ |
| EBL_400 | 携带pVio质粒的大肠杆菌BL21(DE3)E. coli BL21(DE3)with plasmid pVio | 本研究构建This study |
| EBL_401-439 | 携带相应质粒的大肠杆菌BL21(DE3)E. coli BL21(DE3)with corresponding plasmids | 本研究构建This study |
Table 2 Strains in this study
| 菌株Strain | 描述Description | 来源 Source |
|---|---|---|
| E. coli JM109 | 用于构建质粒Construct plasmids | 实验室保存Lab stored |
| E. coli BL21(DE3) | 用于生物传感器宿主菌The host of the biosensor | 实验室保存Lab stored |
| E. coli B8 | 敲除trpR tnaA pheA的E. coli BL21(DE3)E. coli BL21(DE3)ΔtrpR ΔtnaA ΔpheA | [ |
| EBL_400 | 携带pVio质粒的大肠杆菌BL21(DE3)E. coli BL21(DE3)with plasmid pVio | 本研究构建This study |
| EBL_401-439 | 携带相应质粒的大肠杆菌BL21(DE3)E. coli BL21(DE3)with corresponding plasmids | 本研究构建This study |
| 引物Primer | 序列 Sequence(5'-3') | 用途Purpose |
|---|---|---|
| 401_F | ACTTGGAAGGGTAAACTAAGCTGACCGCCAGCTTCG | 扩增pVio以构建pBL_401 Amplifying pVio for pBL_401 |
| 401_R | CTTAGTTTACCCTTCCAAGTTTGTACCAAACGT | |
| 403_F | GTATAAGAGAGAATTTTAATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_403 Amplifying pBL_401 for pBL_403 |
| 403_R | TTGTCATTAAAATTCTCTCTTATACTTAACTAATATACTAAGATGGG | |
| 404_F | GCAGGAAGTAAGTATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_404 Amplifying pBL_401 for pBL_404 |
| 404_R | TTGTCATACTTACTTCCTGCTTATACTTAACTAATATACTAAGATGGG | |
| 405_F | TAGTTAAGTATAAGCGGGGGCTCGAGATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_405 Amplifying pBL_401 for pBL_405 |
| 405_R | CCCCCGCTTATACTTAACTAATATACTAAGATGGG | |
| 406_F | TCGACACACATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_406 Amplifying pBL_401 for pBL_406 |
| 406_R | TAATTTGTCATGTGTGTCGAAACCCCTTATACTTAACTAATATACTAAGATGGG | |
| 407_F | AGAACCGAGAATTATCTCATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_407 Amplifying pBL_401 for pBL_407 |
| 407_R | ATGAGATAATTCTCGGTTCTTATACTTAACTAATATACTAAGATGGG | |
| 415V_F | ATAAGAAGGAGATATACATAATGAGCATTCTCGACTTTCC | 扩增pBL_401以构建pBL_415 Amplifying pBL_401 for pBL_415 |
| 415V_R | GTCATGGTATATCTCCTTATTAAAGTTAAACAAAATTATT | |
| T7terA_F | GAGCGCGCATGAGGCAGCAGCCATCAC | 扩增pBL_401的T7终止子序列以构建pBL_415 Amplifying the T7 terminator sequence of pBL_401 for pBL_415 |
| T7terA_R | CCGTTTAGAGGCCCCAAGGGGTTATGCTAGATACGATTACTTTCTGTTCGACT | |
| T7terB_F | CCCTTGGGGCCTCTAAACGGGTCTTGAGGGGTTTTTTGTGTACACGGCCGCAT | |
| T7terB_R | TATGTATATCTCCTTCTTATACTTAACTAATATACTAAGA | |
| 415_VioA_F | TAAGGAGATATACCATGACAAATTATTCTGACATTTGCAT | 扩增pBL_401的Dug.B2_vioA序列以构建pBL_415 Amplifying Dug.B2_vioA sequence of pBL_401 for pBL_415 |
| 415_VioA_R | CTGCTGCCTCATGCGCGCTCAGTAAGA | |
| 416-420V_F | GGCAGCAGCCATCACCAT | 扩增pBL_415以作为pBL_416-420的载体 Amplifying pBL_415 as their vectors for pBL_416-420 |
| 416-420V_R | GGTATATCTCCTTATTAAAGTTAAACAAAATTATTTCTACAGGGG | |
| 416_VioA_F | TTAACTTTAATAAGGAGATATACCATGAGCACCTATAGCGATATTTGCATTG | 扩增Jan_vioA以构建pBL_416 Amplifying Jan_vioA for pBL_416 |
| 416_VioA_R | ATGGTGATGGCTGCTGCCTTACGCGCGTTCGGTGC | |
| 417_VioA_F | TTAACTTTAATAAGGAGATATACCATGACCAACTATAGCGATATTTGCATTATTGGC | 扩增Pse_vioA以构建pBL_417 Amplifying Pse_vioA for pBL_417 |
| 417_VioA_R | ATGGTGATGGCTGCTGCCTTACGCGCTTTCCGCCG | |
| 418_VioA_F | TTAACTTTAATAAGGAGATATACCATGACCCATCATAGCGATATTTGTATTGTTGG | 扩增Myx_vioA以构建pBL_418 Amplifying Myx_vioA for pBL_418 |
| 418_VioA_R | ATGGTGATGGCTGCTGCCTTACGGGGTGCGATACGCAAT | |
| 419_VioA_F | TTAACTTTAATAAGGAGATATACCATGAAACATAGCAGCGATATTTGCATTGT | 扩增Chr_vioA以构建pBL_419 Amplifying Chr_vioA for pBL_419 |
| 419_VioA_R | ATGGTGATGGCTGCTGCCTTACGCCGCAATGCGCTG | |
| 420_VioA_F | TTAACTTTAATAAGGAGATATACCATGGGCCGCCGCTTTC | 扩增Dug.HH105_vioA以构建pBL_420 Amplifying Dug.HH105_vioA for pBL_420 |
| 420_VioA_R | ATGGTGATGGCTGCTGCCTTACACGCGTTCCGCGGT | |
| 421_F | AGTATAAGGCATCGTAGAAGGGATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_421 Amplifying pBL_401 for pBL_421 |
| 421_R | CTTCTACGATGCCTTATACTTAACTAATATACTAAGATGGG | |
| 422_F | TATAAGACCTGGGACACAAATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_422 Amplifying pBL_401 for pBL_422 |
| 422_R | TTTGTGTCCCAGGTCTTATACTTAACTAATATACTAAGATGGG | |
| 423_F | CTAAACCCTTTTGCAAAATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_423 Amplifying pBL_401 for pBL_423 |
| 423_R | CATTTTGCAAAAGGGTTTAGCTTATACTTAACTAATATACTAAGATGG | |
| 425_F | CTGTAGAAATAATTTTGTTTAACTTTAATAAGGGGTCCAGAACATGACAAATTATTCTGACATTTGCATAGTT | 扩增pBL_415以构建pBL_425 Amplifying pBL_415 for pBL_425 |
| 429_F | CTGTAGAAATAATTTTGTTTAACTTTAATAGGTAGGGGGACATGAGCACCTATAGCGATATTTGC | 扩增pBL_416以构建pBL_429 Amplifying pBL_416 for pBL_429 |
| 437_F | CTGTAGAAATAATTTTGTTTAACTTTAATTCGGAGAGAAAACACATGAAACATAGCAGCGATATTTGCA | 扩增pBL_419以构建pBL_437 Amplifying pBL_419 for pBL_437 |
| 438_F | CTGTAGAAATAATTTTGTTTAACTTTAATAGGGTAAACACATGAAACATAGCAGCGATATTTGCA | 扩增pBL_419以构建pBL_438 Amplifying pBL_419 for pBL_438 |
| 439_F | CTGTAGAAATAATTTTGTTTAACTTTAATTACAGTATTGACATGAAACATAGCAGCGATATTTGCA | 扩增pBL_419以构建pBL_439 Amplifying pBL_419 for pBL_439 |
| adj_RBSAll_R | ATTAAAGTTAAACAAAATTATTTCTACAGGGGAATT | 425_F, 429_F, 437_F, 438_F和439_F的反向引物 The reverse primer for 425_F, 429_F, 437_F, 438_F and 439_F |
Table 3 Primers in this study
| 引物Primer | 序列 Sequence(5'-3') | 用途Purpose |
|---|---|---|
| 401_F | ACTTGGAAGGGTAAACTAAGCTGACCGCCAGCTTCG | 扩增pVio以构建pBL_401 Amplifying pVio for pBL_401 |
| 401_R | CTTAGTTTACCCTTCCAAGTTTGTACCAAACGT | |
| 403_F | GTATAAGAGAGAATTTTAATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_403 Amplifying pBL_401 for pBL_403 |
| 403_R | TTGTCATTAAAATTCTCTCTTATACTTAACTAATATACTAAGATGGG | |
| 404_F | GCAGGAAGTAAGTATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_404 Amplifying pBL_401 for pBL_404 |
| 404_R | TTGTCATACTTACTTCCTGCTTATACTTAACTAATATACTAAGATGGG | |
| 405_F | TAGTTAAGTATAAGCGGGGGCTCGAGATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_405 Amplifying pBL_401 for pBL_405 |
| 405_R | CCCCCGCTTATACTTAACTAATATACTAAGATGGG | |
| 406_F | TCGACACACATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_406 Amplifying pBL_401 for pBL_406 |
| 406_R | TAATTTGTCATGTGTGTCGAAACCCCTTATACTTAACTAATATACTAAGATGGG | |
| 407_F | AGAACCGAGAATTATCTCATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_407 Amplifying pBL_401 for pBL_407 |
| 407_R | ATGAGATAATTCTCGGTTCTTATACTTAACTAATATACTAAGATGGG | |
| 415V_F | ATAAGAAGGAGATATACATAATGAGCATTCTCGACTTTCC | 扩增pBL_401以构建pBL_415 Amplifying pBL_401 for pBL_415 |
| 415V_R | GTCATGGTATATCTCCTTATTAAAGTTAAACAAAATTATT | |
| T7terA_F | GAGCGCGCATGAGGCAGCAGCCATCAC | 扩增pBL_401的T7终止子序列以构建pBL_415 Amplifying the T7 terminator sequence of pBL_401 for pBL_415 |
| T7terA_R | CCGTTTAGAGGCCCCAAGGGGTTATGCTAGATACGATTACTTTCTGTTCGACT | |
| T7terB_F | CCCTTGGGGCCTCTAAACGGGTCTTGAGGGGTTTTTTGTGTACACGGCCGCAT | |
| T7terB_R | TATGTATATCTCCTTCTTATACTTAACTAATATACTAAGA | |
| 415_VioA_F | TAAGGAGATATACCATGACAAATTATTCTGACATTTGCAT | 扩增pBL_401的Dug.B2_vioA序列以构建pBL_415 Amplifying Dug.B2_vioA sequence of pBL_401 for pBL_415 |
| 415_VioA_R | CTGCTGCCTCATGCGCGCTCAGTAAGA | |
| 416-420V_F | GGCAGCAGCCATCACCAT | 扩增pBL_415以作为pBL_416-420的载体 Amplifying pBL_415 as their vectors for pBL_416-420 |
| 416-420V_R | GGTATATCTCCTTATTAAAGTTAAACAAAATTATTTCTACAGGGG | |
| 416_VioA_F | TTAACTTTAATAAGGAGATATACCATGAGCACCTATAGCGATATTTGCATTG | 扩增Jan_vioA以构建pBL_416 Amplifying Jan_vioA for pBL_416 |
| 416_VioA_R | ATGGTGATGGCTGCTGCCTTACGCGCGTTCGGTGC | |
| 417_VioA_F | TTAACTTTAATAAGGAGATATACCATGACCAACTATAGCGATATTTGCATTATTGGC | 扩增Pse_vioA以构建pBL_417 Amplifying Pse_vioA for pBL_417 |
| 417_VioA_R | ATGGTGATGGCTGCTGCCTTACGCGCTTTCCGCCG | |
| 418_VioA_F | TTAACTTTAATAAGGAGATATACCATGACCCATCATAGCGATATTTGTATTGTTGG | 扩增Myx_vioA以构建pBL_418 Amplifying Myx_vioA for pBL_418 |
| 418_VioA_R | ATGGTGATGGCTGCTGCCTTACGGGGTGCGATACGCAAT | |
| 419_VioA_F | TTAACTTTAATAAGGAGATATACCATGAAACATAGCAGCGATATTTGCATTGT | 扩增Chr_vioA以构建pBL_419 Amplifying Chr_vioA for pBL_419 |
| 419_VioA_R | ATGGTGATGGCTGCTGCCTTACGCCGCAATGCGCTG | |
| 420_VioA_F | TTAACTTTAATAAGGAGATATACCATGGGCCGCCGCTTTC | 扩增Dug.HH105_vioA以构建pBL_420 Amplifying Dug.HH105_vioA for pBL_420 |
| 420_VioA_R | ATGGTGATGGCTGCTGCCTTACACGCGTTCCGCGGT | |
| 421_F | AGTATAAGGCATCGTAGAAGGGATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_421 Amplifying pBL_401 for pBL_421 |
| 421_R | CTTCTACGATGCCTTATACTTAACTAATATACTAAGATGGG | |
| 422_F | TATAAGACCTGGGACACAAATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_422 Amplifying pBL_401 for pBL_422 |
| 422_R | TTTGTGTCCCAGGTCTTATACTTAACTAATATACTAAGATGGG | |
| 423_F | CTAAACCCTTTTGCAAAATGACAAATTATTCTGACATTTG | 扩增pBL_401以构建pBL_423 Amplifying pBL_401 for pBL_423 |
| 423_R | CATTTTGCAAAAGGGTTTAGCTTATACTTAACTAATATACTAAGATGG | |
| 425_F | CTGTAGAAATAATTTTGTTTAACTTTAATAAGGGGTCCAGAACATGACAAATTATTCTGACATTTGCATAGTT | 扩增pBL_415以构建pBL_425 Amplifying pBL_415 for pBL_425 |
| 429_F | CTGTAGAAATAATTTTGTTTAACTTTAATAGGTAGGGGGACATGAGCACCTATAGCGATATTTGC | 扩增pBL_416以构建pBL_429 Amplifying pBL_416 for pBL_429 |
| 437_F | CTGTAGAAATAATTTTGTTTAACTTTAATTCGGAGAGAAAACACATGAAACATAGCAGCGATATTTGCA | 扩增pBL_419以构建pBL_437 Amplifying pBL_419 for pBL_437 |
| 438_F | CTGTAGAAATAATTTTGTTTAACTTTAATAGGGTAAACACATGAAACATAGCAGCGATATTTGCA | 扩增pBL_419以构建pBL_438 Amplifying pBL_419 for pBL_438 |
| 439_F | CTGTAGAAATAATTTTGTTTAACTTTAATTACAGTATTGACATGAAACATAGCAGCGATATTTGCA | 扩增pBL_419以构建pBL_439 Amplifying pBL_419 for pBL_439 |
| adj_RBSAll_R | ATTAAAGTTAAACAAAATTATTTCTACAGGGGAATT | 425_F, 429_F, 437_F, 438_F和439_F的反向引物 The reverse primer for 425_F, 429_F, 437_F, 438_F and 439_F |
| 质粒Plasmids | RBSa/RBS | 翻译起始速率TIR | RBSb | 翻译起始速率TIR |
|---|---|---|---|---|
| pBL_401 | AAGGAGATATACC | / | AAGGAGATATACATA | 11 602.81 |
| pBL_403 | AAGGAGATATACC | / | AGAGAATTTTA | 23 837.83 |
| pBL_404 | AAGGAGATATACC | / | CAGGAAGTAAGT | 17 039.41 |
| pBL_405 | AAGGAGATATACC | / | CGGGGGCTCGAG | 7 953.16 |
| pBL_406 | AAGGAGATATACC | / | GGGTTTCGACACAC | 4 941.17 |
| pBL_407 | AAGGAGATATACC | / | AACCGAGAATTATCTC | 1 875.68 |
| pBL_415 | AAGGAGATATACC | 40 994.72 | AAGGAGATATACATA | / |
| pBL_416 | AAGGAGATATACC | 15 231.17 | AAGGAGATATACATA | / |
| pBL_417 | AAGGAGATATACC | 51 571.35 | AAGGAGATATACATA | / |
| pBL_418 | AAGGAGATATACC | 28 729.01 | AAGGAGATATACATA | / |
| pBL_419 | AAGGAGATATACC | 94 682.27 | AAGGAGATATACATA | / |
| pBL_420 | AAGGAGATATACC | 18 153.47 | AAGGAGATATACATA | / |
| pBL_421 | AAGGAGATATA | / | GCATCGTAGAAGGG | 998.41 |
| pBL_422 | AAGGAGATATA | / | ACCTGGGACACAA | 510.31 |
| pBL_423 | AAGGAGATATA | / | CTAAACCCTTTTGCAAA | 99.95 |
| pBL_425 | AAGGGGTCCAGAAC | 10 027.16 | AAGGAGATATACATA | / |
| pBL_429 | AGGTAGGGGGAC | 5 184.17 | AAGGAGATATACATA | / |
| pBL_437 | TCGGAGAGAAAACAC | 10 446.34 | AAGGAGATATACATA | / |
| pBL_438 | AGGGTAAACAC | 4 924.30 | AAGGAGATATACATA | / |
| pBL_439 | TACAGTATTGAC | 2 004.59 | AAGGAGATATACATA | / |
| pED-RBS1 | AAAAAAAGTTTTG | 995.63 | / | / |
| pED-RBS2 | GAAAAAACTCAGC | 393.13 | / | / |
| pED-RBS3 | AAGGAGACCAGC | 12 922.15 | / | / |
| pED-RBS4 | AAAAGGAAAATAA | 7 438.00 | / | / |
Table 4 RBS sequence and its TIR designed in this study
| 质粒Plasmids | RBSa/RBS | 翻译起始速率TIR | RBSb | 翻译起始速率TIR |
|---|---|---|---|---|
| pBL_401 | AAGGAGATATACC | / | AAGGAGATATACATA | 11 602.81 |
| pBL_403 | AAGGAGATATACC | / | AGAGAATTTTA | 23 837.83 |
| pBL_404 | AAGGAGATATACC | / | CAGGAAGTAAGT | 17 039.41 |
| pBL_405 | AAGGAGATATACC | / | CGGGGGCTCGAG | 7 953.16 |
| pBL_406 | AAGGAGATATACC | / | GGGTTTCGACACAC | 4 941.17 |
| pBL_407 | AAGGAGATATACC | / | AACCGAGAATTATCTC | 1 875.68 |
| pBL_415 | AAGGAGATATACC | 40 994.72 | AAGGAGATATACATA | / |
| pBL_416 | AAGGAGATATACC | 15 231.17 | AAGGAGATATACATA | / |
| pBL_417 | AAGGAGATATACC | 51 571.35 | AAGGAGATATACATA | / |
| pBL_418 | AAGGAGATATACC | 28 729.01 | AAGGAGATATACATA | / |
| pBL_419 | AAGGAGATATACC | 94 682.27 | AAGGAGATATACATA | / |
| pBL_420 | AAGGAGATATACC | 18 153.47 | AAGGAGATATACATA | / |
| pBL_421 | AAGGAGATATA | / | GCATCGTAGAAGGG | 998.41 |
| pBL_422 | AAGGAGATATA | / | ACCTGGGACACAA | 510.31 |
| pBL_423 | AAGGAGATATA | / | CTAAACCCTTTTGCAAA | 99.95 |
| pBL_425 | AAGGGGTCCAGAAC | 10 027.16 | AAGGAGATATACATA | / |
| pBL_429 | AGGTAGGGGGAC | 5 184.17 | AAGGAGATATACATA | / |
| pBL_437 | TCGGAGAGAAAACAC | 10 446.34 | AAGGAGATATACATA | / |
| pBL_438 | AGGGTAAACAC | 4 924.30 | AAGGAGATATACATA | / |
| pBL_439 | TACAGTATTGAC | 2 004.59 | AAGGAGATATACATA | / |
| pED-RBS1 | AAAAAAAGTTTTG | 995.63 | / | / |
| pED-RBS2 | GAAAAAACTCAGC | 393.13 | / | / |
| pED-RBS3 | AAGGAGACCAGC | 12 922.15 | / | / |
| pED-RBS4 | AAAAGGAAAATAA | 7 438.00 | / | / |
| 比对蛋白Comparative protein | 相似度Identity/% | |
|---|---|---|
| Dug.B2_VioA | Jan_VioA | 62.5 |
| Pse_VioA | 62.0 | |
| Myx_VioA | 57.9 | |
| Chr_VioA | 54.7 | |
| Dug.HH105_VioA | 59.5 | |
| Jan_VioA | Pse_VioA | 82.5 |
| Myx_VioA | 74.3 | |
| Chr_VioA | 66.4 | |
| Dug.HH105_VioA | 81.4 | |
| Pse_VioA | Myx_VioA | 73.1 |
| Chr_VioA | 65.8 | |
| Dug.HH105_VioA | 80.4 | |
| Myx_VioA | Chr_VioA | 67.1 |
| Dug.HH105_VioA | 73.8 | |
| Chr_VioA | Dug.HH105_VioA | 64.9 |
Table 5 Sequence comparison of 5 VioAs from different sources
| 比对蛋白Comparative protein | 相似度Identity/% | |
|---|---|---|
| Dug.B2_VioA | Jan_VioA | 62.5 |
| Pse_VioA | 62.0 | |
| Myx_VioA | 57.9 | |
| Chr_VioA | 54.7 | |
| Dug.HH105_VioA | 59.5 | |
| Jan_VioA | Pse_VioA | 82.5 |
| Myx_VioA | 74.3 | |
| Chr_VioA | 66.4 | |
| Dug.HH105_VioA | 81.4 | |
| Pse_VioA | Myx_VioA | 73.1 |
| Chr_VioA | 65.8 | |
| Dug.HH105_VioA | 80.4 | |
| Myx_VioA | Chr_VioA | 67.1 |
| Dug.HH105_VioA | 73.8 | |
| Chr_VioA | Dug.HH105_VioA | 64.9 |
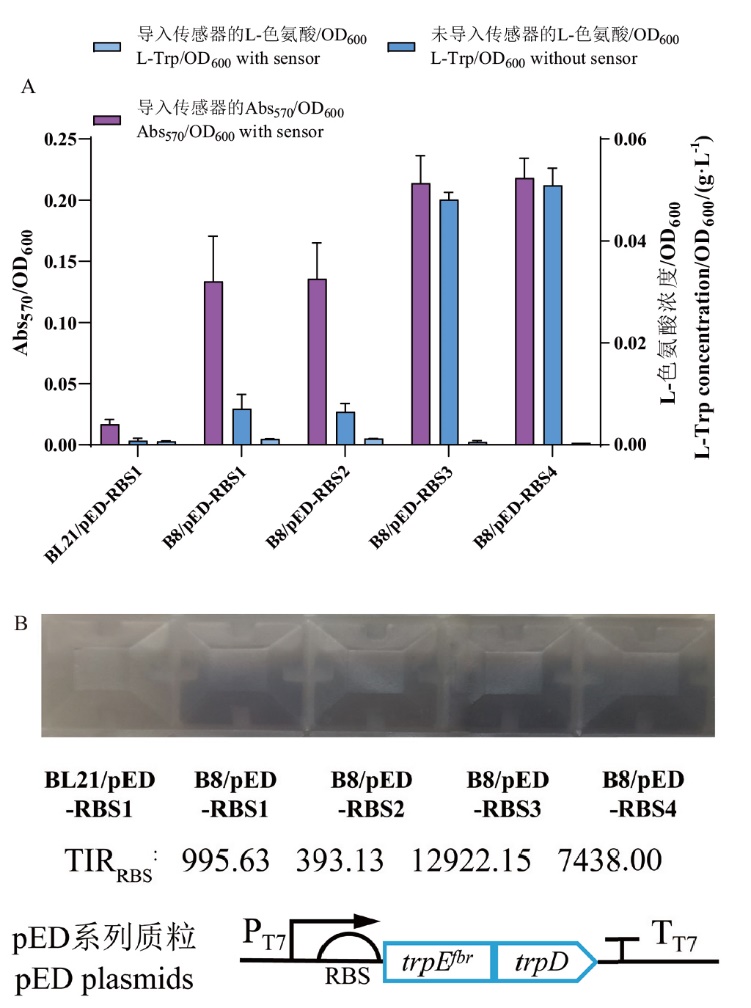
Fig. 8 Testing of the biosensor pBL_439 in different E. coli strains with varying L-Trp production A: Correlation test between biosensor pBL_439 response values and L-Trp production. B: Observational photos by naked eye and schematic diagram of pED series plasmids
| [1] |
Leuchtenberger W, Huthmacher K, Drauz K. Biotechnological production of amino acids and derivatives: current status and prospects[J]. Appl Microbiol Biotechnol, 2005, 69(1): 1-8.
doi: 10.1007/s00253-005-0155-y pmid: 16195792 |
| [2] |
Azuma S, Tsunekawa H, Okabe M, et al. Hyper-production of l-trytophan via fermentation with crystallization[J]. Appl Microbiol Biotechnol, 1993, 39(4): 471-476.
doi: 10.1007/BF00205035 URL |
| [3] |
Guo DY, Kong SJ, Chu X, et al. De novo biosynthesis of indole-3-acetic acid in engineered Escherichia coli[J]. J Agric Food Chem, 2019, 67(29): 8186-8190.
doi: 10.1021/acs.jafc.9b02048 URL |
| [4] |
Wang HJ, Liu WQ, Shi F, et al. Metabolic pathway engineering for high-level production of 5-hydroxytryptophan in Escherichia coli[J]. Metab Eng, 2018, 48: 279-287.
doi: 10.1016/j.ymben.2018.06.007 URL |
| [5] |
Liu S, Xu JZ, Zhang WG. Advances and prospects in metabolic engineering of Escherichia coli for L-tryptophan production[J]. World J Microbiol Biotechnol, 2022, 38(2): 22.
doi: 10.1007/s11274-021-03212-1 |
| [6] |
Wang JL, Ma WJ, Zhou JW, et al. Microbial chassis design and engineering for production of amino acids used in food industry[J]. Syst Microbiol and Biomanuf, 2023, 3(1): 28-48.
doi: 10.1007/s43393-022-00137-0 |
| [7] |
Chen YY, Liu YF, Ding DQ, et al. Rational design and analysis of an Escherichia coli strain for high-efficiency tryptophan production[J]. J Ind Microbiol Biotechnol, 2018, 45(5): 357-367.
doi: 10.1007/s10295-018-2020-x URL |
| [8] |
Gu PF, Yang F, Kang JH, et al. One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli[J]. Microb Cell Fact, 2012, 11: 30.
doi: 10.1186/1475-2859-11-30 |
| [9] |
Ding DQ, Bai DY, Li JL, et al. Analyzing the genetic characteristics of a tryptophan-overproducing Escherichia coli[J]. Bioprocess Biosyst Eng, 2021, 44(8): 1685-1697.
doi: 10.1007/s00449-021-02552-4 |
| [10] |
Liu LN, Duan XG, Wu J. L-tryptophan production in Escherichia coli improved by weakening the pta-AckA pathway[J]. PLoS One, 2016, 11(6): e0158200.
doi: 10.1371/journal.pone.0158200 URL |
| [11] | Guo L, Ding S, Liu YD, et al. Enhancing tryptophan production by balancing precursors in Escherichia coli[J]. Biotech & Bioengineering, 2022, 119(3): 983-993. |
| [12] |
Du LH, Zhang Z, Xu QY, et al. Central metabolic pathway modification to improve L-tryptophan production in Escherichia coli[J]. Bioengineered, 2019, 10(1): 59-70.
doi: 10.1080/21655979.2019.1592417 URL |
| [13] |
Li Y, Mensah EO, Fordjour E, et al. Recent advances in high-throughput metabolic engineering: generation of oligonucleotide-mediated genetic libraries[J]. Biotechnol Adv, 2022, 59: 107970.
doi: 10.1016/j.biotechadv.2022.107970 URL |
| [14] |
Skjoedt ML, Snoek T, Kildegaard KR, et al. Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast[J]. Nat Chem Biol, 2016, 12(11): 951-958.
doi: 10.1038/nchembio.2177 pmid: 27642864 |
| [15] |
D’Oelsnitz S, Nguyen V, Alper HS, et al. Evolving a generalist biosensor for bicyclic monoterpenes[J]. ACS Synth Biol, 2022, 11(1): 265-272.
doi: 10.1021/acssynbio.1c00402 pmid: 34985281 |
| [16] |
Liu YF, Yuan HL, Ding DQ, et al. Establishment of a biosensor-based high-throughput screening platform for tryptophan overproduction[J]. ACS Synth Biol, 2021, 10(6): 1373-1383.
doi: 10.1021/acssynbio.0c00647 pmid: 34081459 |
| [17] |
Fang MY, Wang TM, Zhang C, et al. Intermediate-sensor assisted push-pull strategy and its application in heterologous deoxyviolacein production in Escherichia coli[J]. Metab Eng, 2016, 33: 41-51.
doi: 10.1016/j.ymben.2015.10.006 URL |
| [18] |
Ahmed A, Ahmad A, Li RH, et al. Recent advances in synthetic, industrial and biological applications of violacein and its heterologous production[J]. J Microbiol Biotechnol, 2021, 31(11): 1465-1480.
doi: 10.4014/jmb.2107.07045 URL |
| [19] |
Balibar CJ, Walsh CT. In vitro biosynthesis of violacein from l-tryptophan by the enzymes VioA-E from Chromobacterium violaceum[J]. Biochemistry, 2006, 45(51): 15444-15457.
pmid: 17176066 |
| [20] | Yang D, Park S, Lee S. Production of rainbow colorants by metabolically engineered Escherichia coli[J]. Advanced Science, 2021, 8(13): e2100743. |
| [21] |
Durán N, Justo GZ, Durán M, et al. Advances in Chromobacterium violaceum and properties of violacein-Its main secondary metabolite: a review[J]. Biotechnol Adv, 2016, 34(5): 1030-1045.
doi: 10.1016/j.biotechadv.2016.06.003 URL |
| [22] |
Kordi M, Salami R, Bolouri P, et al. White biotechnology and the production of bio-products[J]. Syst Microbiol and Biomanuf, 2022, 2(3): 413-429.
doi: 10.1007/s43393-022-00078-8 |
| [23] |
Mendes AS, de Carvalho JE, Duarte MCT, et al. Factorial design and response surface optimization of crude violacein for Chromobacterium violaceum production[J]. Biotechnol Lett, 2001, 23(23): 1963-1969.
doi: 10.1023/A:1013734315525 URL |
| [24] |
Wang HS, Jiang PX, Lu Y, et al. Optimization of culture conditions for violacein production by a new strain of Duganella sp. B2[J]. Biochem Eng J, 2009, 44(2/3): 119-124.
doi: 10.1016/j.bej.2008.11.008 URL |
| [25] |
Yang C, Jiang PX, Xiao S, et al. Fed-batch fermentation of recombinant Citrobacter freundii with expression of a violacein-synthesizing gene cluster for efficient violacein production from glycerol[J]. Biochem Eng J, 2011, 57: 55-62.
doi: 10.1016/j.bej.2011.08.008 URL |
| [26] |
Fang MY, Zhang C, Yang S, et al. High crude violacein production from glucose by Escherichia coli engineered with interactive control of tryptophan pathway and violacein biosynthetic pathway[J]. Microb Cell Factories, 2015, 14(1): 1-13.
doi: 10.1186/s12934-014-0183-3 URL |
| [27] |
Liu J, Xu JZ, Rao ZM, et al. An enzymatic colorimetric whole-cell biosensor for high-throughput identification of lysine overproducers[J]. Biosens Bioelectron, 2022, 216: 114681.
doi: 10.1016/j.bios.2022.114681 URL |
| [28] |
Yang D, Kim WJ, Yoo SM, et al. Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria[J]. Proc Natl Acad Sci USA, 2018, 115(40): 9835-9844.
doi: 10.1073/pnas.1808567115 pmid: 30232266 |
| [29] |
Hui CY, Guo Y, Zhu DL, et al. Metabolic engineering of the violacein biosynthetic pathway toward a low-cost, minimal-equipment lead biosensor[J]. Biosens Bioelectron, 2022, 214: 114531.
doi: 10.1016/j.bios.2022.114531 URL |
| [30] |
Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression[J]. Nat Biotechnol, 2009, 27(10): 946-950.
doi: 10.1038/nbt.1568 pmid: 19801975 |
| [31] |
Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar[J]. Anal Chem, 1959, 31(3): 426-428.
doi: 10.1021/ac60147a030 URL |
| [32] |
Wu HX, Yang JH, Shen PJ, et al. High-level production of indole-3-acetic acid in the metabolically engineered Escherichia coli[J]. J Agric Food Chem, 2021, 69(6): 1916-1924.
doi: 10.1021/acs.jafc.0c08141 URL |
| [33] |
Lin JL, Wagner JM, Alper HS. Enabling tools for high-throughput detection of metabolites: metabolic engineering and directed evolution applications[J]. Biotechnol Adv, 2017, 35(8): 950-970.
doi: 10.1016/j.biotechadv.2017.07.005 URL |
| [34] |
Rogers JK, Taylor ND, Church GM. Biosensor-based engineering of biosynthetic pathways[J]. Curr Opin Biotechnol, 2016, 42: 84-91.
doi: 10.1016/j.copbio.2016.03.005 URL |
| [35] |
Füller JJ, Röpke R, Krausze J, et al. Biosynthesis of violacein, structure and function of l-tryptophan oxidase VioA from Chromobacterium violaceum[J]. J Biol Chem, 2016, 291(38): 20068-20084.
doi: 10.1074/jbc.M116.741561 pmid: 27466367 |
| [36] |
叶健文, 陈江楠, 张旭, 等. 动态调控: 一种高效的细胞工厂工程化代谢改造策略[J]. 生物技术通报, 2020, 36(6):1-12
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0317 |
| Ye JW, Chen JN, Zhang X, et al. Dynamic control: an efficient strategy for metabolically engineering microbial cell factories[J]. Biotechnol Bull, 2020, 36(6):1-12 | |
| [37] |
Ikeda M. Towards bacterial strains overproducing l-tryptophan and other aromatics by metabolic engineering[J]. Appl Microbiol Biotechnol, 2006, 69(6): 615-626.
doi: 10.1007/s00253-005-0252-y pmid: 16374633 |
| [38] |
Alkhalaf L, Ryan K. Biosynthetic manipulation of tryptophan in bacteria: pathways and mechanisms[J]. Chem Biol, 2015, 22(3): 317-328.
doi: 10.1016/j.chembiol.2015.02.005 URL |
| [39] |
Almeida MC, Resende DISP, da Costa PM, et al. Tryptophan derived natural marine alkaloids and synthetic derivatives as promising antimicrobial agents[J]. Eur J Med Chem, 2021, 209: 112945.
doi: 10.1016/j.ejmech.2020.112945 URL |
| [1] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [2] | CHEN Cai-ping, REN Hao, LONG Teng-fei, HE Bing, LU Zhao-xiang, SUN Jian. Research Advances in the Treatment of Inflammation Bowel Disease Using Escherichia coli Nissle 1917 [J]. Biotechnology Bulletin, 2023, 39(6): 109-118. |
| [3] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [4] | TANG Rui-qi, ZHAO Xin-qing, ZHU Du, WANG Ya. Stress Tolerance of Escherichia coli to Inhibitors in Lignocellulosic Hydrolysates [J]. Biotechnology Bulletin, 2023, 39(11): 205-216. |
| [5] | CHEN Xiao-lin, LIU Yang-er, XU Wen-tao, GUO Ming-zhang, LIU Hui-lin. Application of Synthetic Biology Based Whole-cell Biosensor Technology in the Rapid Detection of Food Safety [J]. Biotechnology Bulletin, 2023, 39(1): 137-149. |
| [6] | LI Hai-li, LANG Li-min, ZHANG Qing-xian, YOU Yi, ZHU Wen-hao, WANG Zhi-fang, ZHANG Li-xian, WANG Ke-ling. Identification and Drug Resistance of Escherichia coli Simultaneously Producing Carbapenemase NDM-1 and NDM-5 [J]. Biotechnology Bulletin, 2022, 38(9): 106-115. |
| [7] | CHENG Shen-wei, ZHANG Ke-qiang, LIANG Jun-feng, LIU Fu-yuan, GAO Xing-liang, DU Lian-zhu. Establishment of a Triple Droplet Digital PCR Quantitative Detection Method for Typical Pathogenic Bacteria in Livestock and Poultry Manure [J]. Biotechnology Bulletin, 2022, 38(9): 271-280. |
| [8] | ZHAO Yan-kun, LIU Hui-min, MENG Lu, WANG Cheng, WANG Jia-qi, ZHENG Nan. Research Progress in Heteroresistance of Escherichia coli [J]. Biotechnology Bulletin, 2022, 38(9): 59-71. |
| [9] | GAO Wei-xin, HUANG Huo-qing, ZHAO Jing, ZHANG Xin, YANG Ning, YANG Hao-meng. Construction and Activity Verification of Ribonucleoprotein Complex for Gene Editing [J]. Biotechnology Bulletin, 2022, 38(8): 60-68. |
| [10] | SUN Man-luan, GE Sai, BU Jia, ZHU Zhuang-yan. Regulation Mechanism of Ribonucleases in Escherichia coli [J]. Biotechnology Bulletin, 2022, 38(3): 234-245. |
| [11] | LI Xiao-fang, LIU Hui-yan, PAN Lin, AI Zhi-yu, LI Yi-ming, ZHANG Heng, FANG Hai-tian. Breeding High-yield L-isoleucine Escherichia coli by ARTP Mutagenesis [J]. Biotechnology Bulletin, 2022, 38(1): 150-156. |
| [12] | WU Rong, CAO Jia-rui, CAO Jun, LIU Fei-xiang, YANG Meng, SU Er-zheng. Expression and Fermentation Optimization of Candida antarctica Lipase B in Escherichia coli [J]. Biotechnology Bulletin, 2021, 37(2): 138-148. |
| [13] | WANG Peng-fei, YANG Min, ZHU Long-jiao, XU Wen-tao. Advances in Biosensors Based on Platinum Nanoclusters [J]. Biotechnology Bulletin, 2021, 37(12): 235-242. |
| [14] | WANG Kai-kai, WANG Xiao-lu, SU Xiao-yun, ZHANG Jie. Optimization and Application of Double-plasmid CRISPR-Cas9 System in Escherichia coli [J]. Biotechnology Bulletin, 2021, 37(12): 252-264. |
| [15] | ZHAO Ying, WANG Nan, LU An-xiang, FENG Xiao-yuan, GUO Xiao-jun, LUAN Yun-xia. Application in the Detection of Fungal Toxins by Nucleic Acid Aptamer Lateral Flow Chromatography Analysis Technique [J]. Biotechnology Bulletin, 2020, 36(8): 217-227. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||