








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (12): 311-319.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0702
Previous Articles Next Articles
SHANG Yi-tong1,2( ), YAN Huan-huan1,2, WANG Li-hong1,2, TIAN Xue-qin1,2, XUE Ping-hong1,2, LUO Tao1, HU Zhi-hong1,2(
), YAN Huan-huan1,2, WANG Li-hong1,2, TIAN Xue-qin1,2, XUE Ping-hong1,2, LUO Tao1, HU Zhi-hong1,2( )
)
Received:2023-07-21
Online:2023-12-26
Published:2024-01-11
Contact:
HU Zhi-hong
E-mail:syt15735905986@163.com;huzhihong426@163.com
SHANG Yi-tong, YAN Huan-huan, WANG Li-hong, TIAN Xue-qin, XUE Ping-hong, LUO Tao, HU Zhi-hong. Study on the Function of Phosphomevalonate Kinase in Aspergillus oryzae[J]. Biotechnology Bulletin, 2023, 39(12): 311-319.
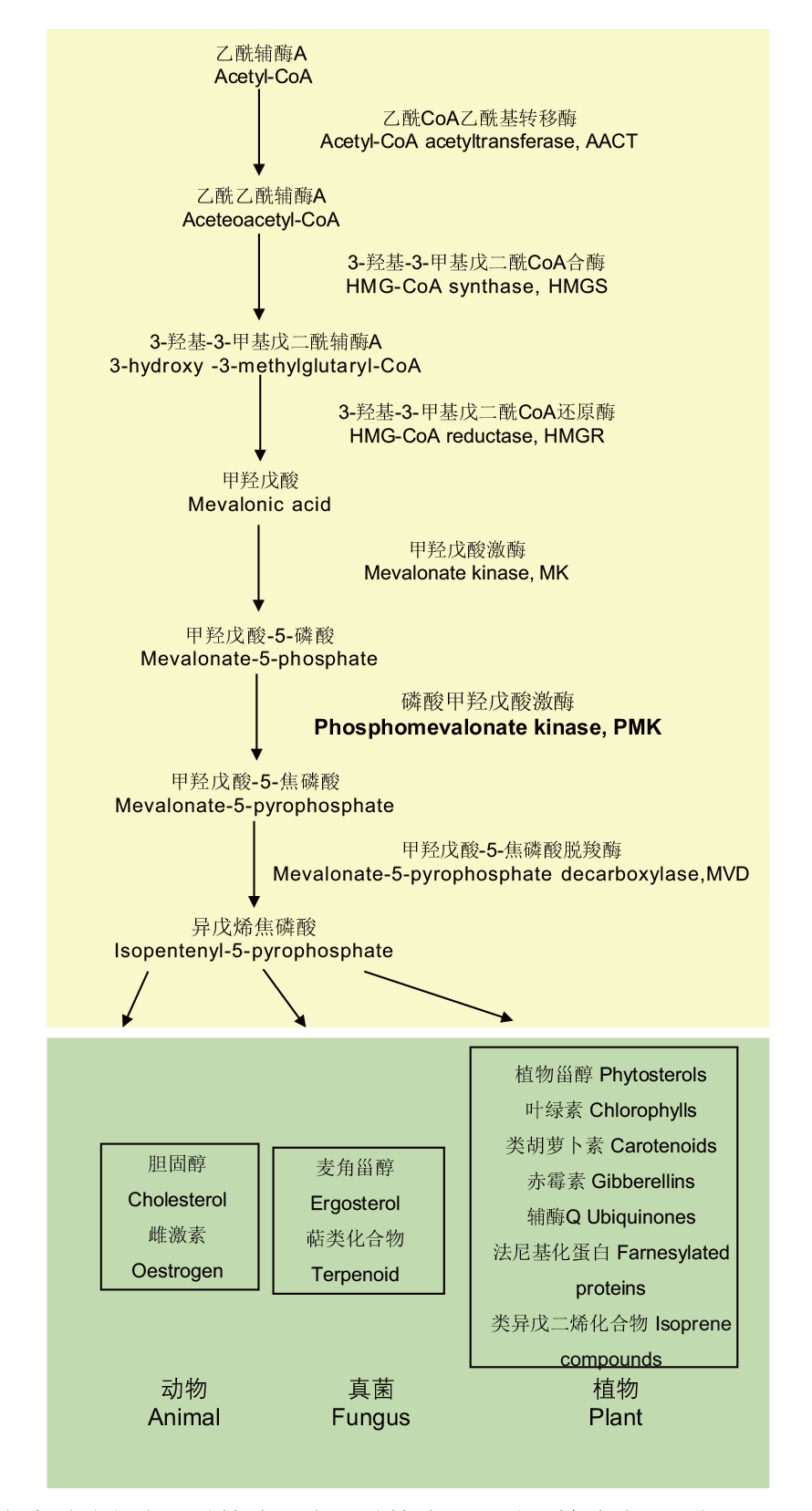
Fig. 1 MVA pathway metabolic synthesis diagram MVA pathway is formed by acetyl-CoA under the catalysis of acetyl-CoA acetyltransferase, 3-hydroxy-3-methylglutaryl-CoA synthase, 3-hydroxy-3-methylglutaryl-CoA reductase, mevalonate kinase, phosphomevalonate kinase, mevalonate 5-pyrophosphate decarboxylase to generate isopentenyl-5-pyrophosphate. In plants, animals,and fungi, it is located in the upstream of isoprenoids and sterols, playing an important role in the growth and metabolic activities of organisms
| 物种名 Species name | 序列号GI No. |
|---|---|
| 黄花蒿Artemisia annua | gi|1387810006 |
| 人参Panax ginseng | gi|555431957 |
| 黄花蒿Artemisia annua | gi|1387770975 |
| 丹参Salvia miltiorrhiza | gi|374639353 |
| 番茄Solanum lycopersicum | gi|1480008164 |
| 海枣Phoenix dactylifera | gi|672109111 |
| 短花药野生稻Oryza brachyantha | gi|1002850285 |
| 玉米Zea mays | gi|1126132679 |
| 卷柏Selaginella moellendorffii | gi|1376963038 |
| 地钱Marchantia polymorpha | gi|1376841057 |
| 小立碗藓Physcomitrium patens | gi|1373905490 |
| 高卢蜜环菌Armillaria gallica | gi|1243522655 |
| 米曲霉Aspergillus oryzae | gi|391866384 |
| 酿酒酵母Saccharomyces cerevisiae | gi|171479 |
| 植物乳杆菌Lactiplantibacillus plantarum | gi|935445216 |
| 金黄色葡萄球菌Staphylococcus aureus | gi|897312969 |
| 欧洲熊蜂Bombus terrestris | gi|526117733 |
| 家蚕Bombyx mori | gi|526117733 |
| 智人Homo sapiens | gi|1294782 |
| 小鼠Mus musculus | gi|32363399 |
Table 1 Erg8 numbers of different species
| 物种名 Species name | 序列号GI No. |
|---|---|
| 黄花蒿Artemisia annua | gi|1387810006 |
| 人参Panax ginseng | gi|555431957 |
| 黄花蒿Artemisia annua | gi|1387770975 |
| 丹参Salvia miltiorrhiza | gi|374639353 |
| 番茄Solanum lycopersicum | gi|1480008164 |
| 海枣Phoenix dactylifera | gi|672109111 |
| 短花药野生稻Oryza brachyantha | gi|1002850285 |
| 玉米Zea mays | gi|1126132679 |
| 卷柏Selaginella moellendorffii | gi|1376963038 |
| 地钱Marchantia polymorpha | gi|1376841057 |
| 小立碗藓Physcomitrium patens | gi|1373905490 |
| 高卢蜜环菌Armillaria gallica | gi|1243522655 |
| 米曲霉Aspergillus oryzae | gi|391866384 |
| 酿酒酵母Saccharomyces cerevisiae | gi|171479 |
| 植物乳杆菌Lactiplantibacillus plantarum | gi|935445216 |
| 金黄色葡萄球菌Staphylococcus aureus | gi|897312969 |
| 欧洲熊蜂Bombus terrestris | gi|526117733 |
| 家蚕Bombyx mori | gi|526117733 |
| 智人Homo sapiens | gi|1294782 |
| 小鼠Mus musculus | gi|32363399 |
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| rH-F | GACAACATCCAGGGTATCACTAAGC |
| rH-R | GGTCTCCTCGTAGATCATGGCA |
| qRT-AoErg8-F | GGCTCTAGTAACTGCCCTAGTA |
| qRT-AoErg8-R | AGCCTGTGCCAAGTTATGAA |
| pEX2B-AoErg8-F | TTCACGTGCCCGTGCTTAAGATGTCTTATCCGCCATCCGGGAG |
| pEX2B-AoErg8-R | GAGGCCATGATATCCTTAAGAAGCCAACCGGCATATTGGTC |
| pYES2.0-AoErg8-F | CTATAGGGAATATTAAGCTTATGTCTTATCCGCCATCCGGG |
| pYES2.0-AoErg8-R | GATGGATATCTGCAGAATTCAAGCCAACCGGCATATTGGTC |
Table 2 Primers used in this study
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| rH-F | GACAACATCCAGGGTATCACTAAGC |
| rH-R | GGTCTCCTCGTAGATCATGGCA |
| qRT-AoErg8-F | GGCTCTAGTAACTGCCCTAGTA |
| qRT-AoErg8-R | AGCCTGTGCCAAGTTATGAA |
| pEX2B-AoErg8-F | TTCACGTGCCCGTGCTTAAGATGTCTTATCCGCCATCCGGGAG |
| pEX2B-AoErg8-R | GAGGCCATGATATCCTTAAGAAGCCAACCGGCATATTGGTC |
| pYES2.0-AoErg8-F | CTATAGGGAATATTAAGCTTATGTCTTATCCGCCATCCGGG |
| pYES2.0-AoErg8-R | GATGGATATCTGCAGAATTCAAGCCAACCGGCATATTGGTC |
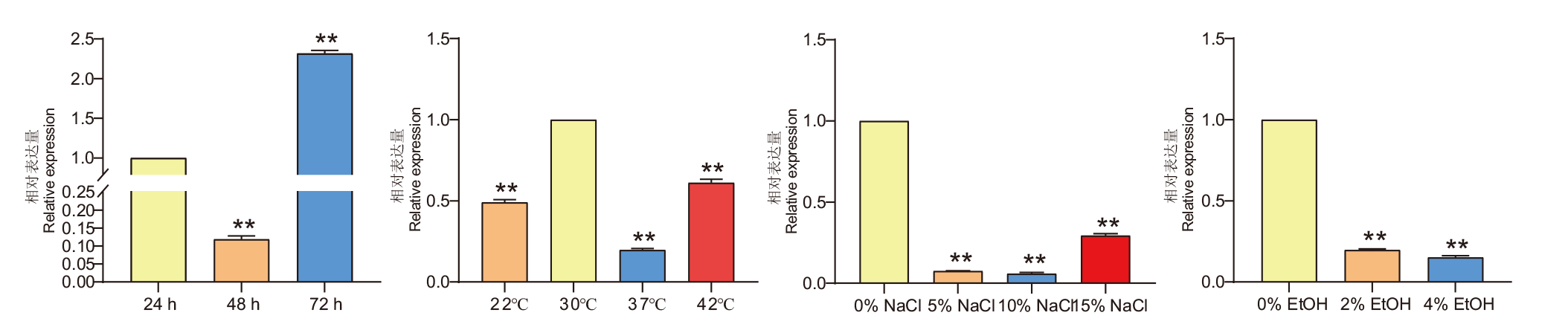
Fig.3 Expressions of AoErg8 on agar CD medium under different growth time and different abiotic stress A: Expression of AoErg8 at 24, 48, and 72 h of growth. B: Expression of AoErg8 at different temperatures. C: Expression of AoErg8 under different salt stress conditions.D: Expression of AoErg8 under different ethanol(EtOH)stress. In order to determine the level of mRNA at different growth times, Mycelium was harvested at 24, 48 and 72 h. For other tests, harvest Mycelium at 72 h. The expression levels of AoErg8 at 24 h, 30℃, and in 0% NaCl/ethanol were used as corresponding references. The numerical value represents the mean ± standard deviation of three independent experiments. Statistical analysis using GraphPad’s t-test, *: P<0.05, **: P <0.01, the same below

Fig. 4 AoErg8 may restore the phenotype of Saccharomyces cerevisiae erg8 mutant A: The growth of wild-type growth, erg8 mutant(Y40835), and AoErg8 transformant on YPD and YPG media at 30℃ and 37℃. B: Measure the ergosterol content in all transformants, incubate the control and transformants in liquid YPG at 30℃ for 2 d, and collect yeast to determine the ergosterol content. Compare the ergosterol content of AoErg8 transformant with the pYES2.0/erg8 mutant in the corresponding culture medium

Fig. 5 Phenotype and ergosterol content of AoErg8 overexpressing strain A: After 72 h of cultivation, the colony morphology of CK(pEX2B vector transformed by wild-type A. oryzae)and AoErg8 overexpressing strains on PDA medium. B: Spore count of different transgenic strains. C: Colony diameter of different transgenic strains. D: Ergosterol content of AoErg8 overexpressing strains. The control and transformants were cultured in DPY at 30℃ for 3 d and collected for determination of ergosterol content. Compare each experimental group with the corresponding control group
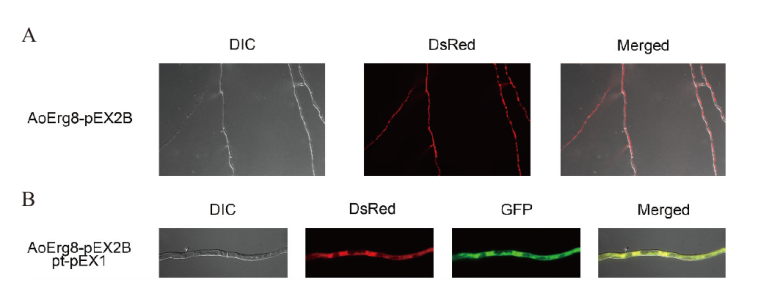
Fig. 6 Subcellular localization of AoErg8 A: The mycelium of A. oryzae 3.042 ΔpyrG transformed with AoErg8-DsRed. Left to right: bright field(DIC), fluorescent images of DsRed and merged image of DsRed and bright field. B: Co-localization of AoErg8 DsRed with cytoplasm. The A. oryzae 3.042 ΔpyrG mycelium co-transformed of AoErg8-DsRed and pt-pEX1-GFP carriers. From left to right: bright field(DIC), fluorescent images of DsRed, GFP, and merged images of DsRed, GFP, and DIC
| [1] |
Lombard J, Moreira D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life[J]. Mol Biol Evol, 2011, 28(1): 87-99.
doi: 10.1093/molbev/msq177 pmid: 20651049 |
| [2] |
Sun YL, Niu YL, Huang H, et al. Mevalonate diphosphate decarboxylase MVD/Erg19 is required for ergosterol biosynthesis, growth, sporulation and stress tolerance in Aspergillus oryzae[J]. Front Microbiol, 2019, 10: 1074.
doi: 10.3389/fmicb.2019.01074 URL |
| [3] | Huang GC, Cai WX, Xu BJ. Vitamin D2, ergosterol, and vitamin B2 content in commercially dried mushrooms marketed in China and increased vitamin D2 content following UV-C irradiation[J]. Int J Vitam Nutr Res, 2017, 87(5/6): 1-10. |
| [4] |
Karpova NV, Andryushina VA, Stytsenko TS, et al. A search for microscopic fungi with directed hydroxylase activity for the synthesis of steroid drugs[J]. Prikl Biokhim Mikrobiol, 2016, 52(3): 324-332.
pmid: 29509389 |
| [5] |
Kobori M, Yoshida M, Ohnishi-Kameyama M, et al. Ergosterol peroxide from an edible mushroom suppresses inflammatory responses in RAW264.7 macrophages and growth of HT29 colon adenocarcinoma cells[J]. Br J Pharmacol, 2007, 150(2): 209-219.
doi: 10.1038/sj.bjp.0706972 URL |
| [6] |
Kitchawalit S, Kanokmedhakul K, Kanokmedhakul S, et al. A new benzyl ester and ergosterol derivatives from the fungus Gymnoascus reessii[J]. Nat Prod Res, 2014, 28(14): 1045-1051.
doi: 10.1080/14786419.2014.903478 pmid: 24708569 |
| [7] |
Houten SM, Waterham HR. Nonorthologous gene displacement of phosphomevalonate kinase[J]. Mol Genet Metab, 2001, 72(3): 273-276.
pmid: 11243736 |
| [8] |
Andreassi JL, Leyh TS. Molecular functions of conserved aspects of the GHMP kinase family[J]. Biochemistry, 2004, 43(46): 14594-14601.
pmid: 15544330 |
| [9] |
Zhang ZH, Li CH, Wu F, et al. Genomic variations of the mevalonate pathway in porokeratosis[J]. eLife, 2015, 4: e06322.
doi: 10.7554/eLife.06322 URL |
| [10] |
Lange BM, Ghassemian M. Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism[J]. Plant Mol Biol, 2003, 51(6): 925-948.
doi: 10.1023/A:1023005504702 URL |
| [11] |
Sando T, Takaoka C, Mukai Y, et al. Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis[J]. Biosci Biotechnol Biochem, 2008, 72(8): 2049-2060.
doi: 10.1271/bbb.80165 URL |
| [12] |
Ma YM, Yuan LC, Wu B, et al. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza[J]. J Exp Bot, 2012, 63(7): 2809-2823.
doi: 10.1093/jxb/err466 URL |
| [13] | 郑汉, 虞慕瑶, 濮春娟, 等. 香樟甲羟戊酸-5-磷酸激酶基因CcPMK的克隆和表达分析[J]. 中国中药杂志, 2020, 45(1): 78-84. |
| Zheng H, Yu MY, Pu CJ, et al. Cloning and expression analysis of 5-phosphomevalonate kinase gene(CcPMK)in Cinnamomum camphora[J]. China J Chin Mater Med, 2020, 45(1): 78-84. | |
| [14] |
Niu MY, Xiong YP, Yan HF, et al. Cloning and expression analysis of mevalonate kinase and phosphomevalonate kinase genes associated with the MVA pathway in Santalum album[J]. Sci Rep, 2021, 11(1): 16913.
doi: 10.1038/s41598-021-96511-4 |
| [15] |
Tsay YH, Robinson GW. Cloning and characterization of ERG8, an essential gene of Saccharomyces cerevisiae that encodes phosphomevalonate kinase[J]. Mol Cell Biol, 1991, 11(2): 620-631.
doi: 10.1128/mcb.11.2.620-631.1991 pmid: 1846667 |
| [16] |
Wang SX, Liu ZC, Wang XT, et al. Cloning and characterization of a phosphomevalonate kinase gene from Sanghuangporus baumii[J]. Biotechnol Biotechnol Equip, 2021, 35(1): 934-942.
doi: 10.1080/13102818.2021.1938678 URL |
| [17] |
Doun SS, Burgner JW, Briggs SD, et al. Enterococcus faecalis phosphomevalonate kinase[J]. Protein Sci, 2005, 14(5): 1134-1139.
doi: 10.1110/ps.041210405 URL |
| [18] | Dairi T. Studies on biosynthetic genes and enzymes of isoprenoids produced by actinomycetes[J]. Jpn J Antibiot, 2005, 58(1): 87-98. |
| [19] |
Kitamoto K. Cell biology of the Koji mold Aspergillus oryzae[J]. Biosci Biotechnol Biochem, 2015, 79(6): 863-869.
doi: 10.1080/09168451.2015.1023249 URL |
| [20] |
Hu ZH, Li GH, Sun YL, et al. Gene transcription profiling of Aspergillus oryzae 3.042 treated with ergosterol biosynthesis inhibitors[J]. Braz J Microbiol, 2019, 50(1): 43-52.
doi: 10.1007/s42770-018-0026-1 |
| [21] |
Sun YL, Niu YL, He B, et al. A dual selection marker transformation system using Agrobacterium tumefaciens for the industrial Aspergillus oryzae 3.042[J]. J Microbiol Biotechnol, 2019, 29(2): 230-234.
doi: 10.4014/jmb.1811.11027 URL |
| [22] |
Huang H, Niu YL, Jin Q, et al. Identification of six thiolases and their effects on fatty acid and ergosterol biosynthesis in Aspergillus oryzae[J]. Appl Environ Microbiol, 2022, 88(6): e0237221.
doi: 10.1128/aem.02372-21 URL |
| [23] |
Jin Q, Li GH, Qin KH, et al. The expression pattern, subcellular localization and function of three sterol 14α-demethylases in Aspergillus oryzae[J]. Front Genet, 2023, 14: 1009746.
doi: 10.3389/fgene.2023.1009746 URL |
| [24] |
Nguyen KT, Ho QN, Do LTBX, et al. A new and efficient approach for construction of uridine/uracil auxotrophic mutants in the filamentous fungus Aspergillus oryzae using Agrobacterium tumefaciens-mediated transformation[J]. World J Microbiol Biotechnol, 2017, 33(6): 107.
doi: 10.1007/s11274-017-2275-9 URL |
| [25] | 黄慧, 王立, 胡志宏, 等. 培养基组分及培养条件对米曲霉麦角甾醇含量的影响[J]. 江西科技师范大学学报, 2021(6): 87-92. |
| Huang H, Wang L, Hu ZH, et al. Effect of medium composition and culture conditions on ergosterol content in Aspergillus oryzae[J]. J Jiangxi Sci Technol Norm Univ, 2021(6): 87-92. | |
| [26] |
Ma SM, Garcia DE, Redding-Johanson AM, et al. Optimization of a heterologous mevalonate pathway through the use of variant HMG-CoA reductases[J]. Metab Eng, 2011, 13(5): 588-597.
doi: 10.1016/j.ymben.2011.07.001 pmid: 21810477 |
| [27] |
Hu ZH, He B, Ma L, et al. Recent advances in ergosterol biosynthesis and regulation mechanisms in Saccharomyces cerevisiae[J]. Indian J Microbiol, 2017, 57(3): 270-277.
doi: 10.1007/s12088-017-0657-1 URL |
| [28] |
Hu ZH, Huang H, Sun YL, et al. Effects on gene transcription profile and fatty acid composition by genetic modification of mevalonate diphosphate decarboxylase MVD/Erg19 in Aspergillus oryzae[J]. Microorganisms, 2019, 7(9): 342.
doi: 10.3390/microorganisms7090342 URL |
| [29] | 何海. 茯苓磷酸甲羟戊酸激酶基因克隆及功能分析[D]. 武汉: 华中农业大学, 2016. |
| He H. Cloning and characterization of phosphomevalonate kinase gene from Poria cocos[D]. Wuhan: Huazhong Agricultural University, 2016. | |
| [30] | 赵乐, 马利刚, 杨方方, 等. 独行菜磷酸甲羟戊酸激酶LaPMK基因克隆、生物信息学分析及原核表达[J]. 中草药, 2016, 47(17): 3087-3093. |
| Zhao L, Ma LG, Yang FF, et al. Cloning, bioinformatic analysis, and prokaryotic expression of LaPMK gene in Lepidium apetalum[J]. Chin Tradit Herb Drugs, 2016, 47(17): 3087-3093. | |
| [31] |
李辉, 温春秀, 刘灵娣, 等. 紫苏甲羟戊酸-5-磷酸激酶基因PfPMK的克隆与表达分析[J]. 华北农学报, 2022, 37(2): 49-55.
doi: 10.7668/hbnxb.20192609 |
| Li H, Wen CX, Liu LD, et al. Cloning and expression analysis of 5-phosphomevalonate kinase PfPMK gene in Perilla frutescens L[J]. Acta Agric Boreali Sin, 2022, 37(2): 49-55. |
| [1] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [2] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [3] | TENG Meng-xin, XU Ya, HE Jing, WANG Qi, QIAO Fei, LI Jing-yang, LI Xin-guo. Cloning and Prokaryotic Expression Analysis of MaMC6 in Banana [J]. Biotechnology Bulletin, 2023, 39(12): 179-186. |
| [4] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [5] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [6] | SHI Ya-nan, WANG De-pei, WANG Yi-chuan, ZHOU Hao, XUE Xian-li. Effects of msn2 Knock-out on the Growth and Kojic Acid Production of Aspergillus oryzae [J]. Biotechnology Bulletin, 2022, 38(8): 188-197. |
| [7] | YANG Jia-bao, ZHOU Zhi-ming, ZHANG Zhan, FENG Li, SUN Li. Cloning,Expression of Helianthus annuus HaLACS1 Gene and Identification of Its Functional Complementation in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2022, 38(6): 147-156. |
| [8] | HAO Qing-qing, YAO Sheng, LIU Jia-he, CHEN Pei-zhen, ZHANG Meng-yang, JI Kong-shu. Cloning and Expression Analysis of NAC Transcription Factor PmNAC8 in Pinus massoniana [J]. Biotechnology Bulletin, 2022, 38(4): 202-216. |
| [9] | DANG Yuan, LI Wei, MIAO Xiang, XIU Yu, LIN Shan-zhi. Cloning of Oleosin Gene PsOLE4 from Prunus sibirica and Its Regulatory Function Analysis for Oil Accumulation [J]. Biotechnology Bulletin, 2022, 38(11): 151-161. |
| [10] | LUO Ying, TANG Zhi, WANG Fan, LIU Xiao-xia, LUO Xiao-fang, HE Fu-lin. Cloning and Response Analysis to Abiotic Stress of GbR2R3-MYB1 Gene from Ginkgo biloba [J]. Biotechnology Bulletin, 2022, 38(10): 184-194. |
| [11] | SUN Rui-fen, ZHANG Yan-fang, NIU Su-qing, GUO Shu-chun, LI Su-ping, YU Hai-feng, NIE Hui, MOU Ying-nan. Expression Analysis and Functional Verification of the HaACO1 Gene in Sunflower [J]. Biotechnology Bulletin, 2021, 37(9): 114-124. |
| [12] | FAN Ya-peng, RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei. Cloning,Expression and Preliminary Bioinformatics Analysis of the Alkaline Tolerant Gene GhZAT12 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 121-130. |
| [13] | SHAN Cao-mei, YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang. Cloning,and Functional Analysis of Gene OsRAI1 Resistant to Hirschmanniella mucronate in Rice [J]. Biotechnology Bulletin, 2021, 37(7): 146-155. |
| [14] | SUN Xiao-qian, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Gene Cloning,Subcellular Localization and Expression Analyses of FtMYBF Transcription Factor in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2021, 37(3): 10-17. |
| [15] | HAN Zhan-hong, ZONG Yuan-yuan, ZHANG Xue-mei, WANG Bin, PRUSKY Dov, BI Yang. Bioinformatics,Subcellular Localization and Expression Analysis of erg4 in Penicillium expansum [J]. Biotechnology Bulletin, 2021, 37(12): 60-70. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||