








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (12): 60-70.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0141
Previous Articles Next Articles
HAN Zhan-hong1( ), ZONG Yuan-yuan1, ZHANG Xue-mei1, WANG Bin1, PRUSKY Dov2, BI Yang1(
), ZONG Yuan-yuan1, ZHANG Xue-mei1, WANG Bin1, PRUSKY Dov2, BI Yang1( )
)
Received:2021-02-04
Online:2021-12-26
Published:2022-01-19
Contact:
BI Yang
E-mail:hanzhanhong009@163.com;biyang@gsau.edu.cn
HAN Zhan-hong, ZONG Yuan-yuan, ZHANG Xue-mei, WANG Bin, PRUSKY Dov, BI Yang. Bioinformatics,Subcellular Localization and Expression Analysis of erg4 in Penicillium expansum[J]. Biotechnology Bulletin, 2021, 37(12): 60-70.
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | F:ATGGAGGACCAAGTCAAATC |
| R:AAAAAGAGAAATTGCAGTAG | |
| erg4B | F:ATGCCCTCCAAAAAGGACTCTC |
| R:GTAAATTCCAGGAATGATGC | |
| erg4C | F:ATGGATCGTCCCGGCTTCATT |
| R:TTAGAAGACATAAGGAATGA |
Table 1 Primers sequence for the clone of CDS of gene erg4A,erg4B and erg4C
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | F:ATGGAGGACCAAGTCAAATC |
| R:AAAAAGAGAAATTGCAGTAG | |
| erg4B | F:ATGCCCTCCAAAAAGGACTCTC |
| R:GTAAATTCCAGGAATGATGC | |
| erg4C | F:ATGGATCGTCCCGGCTTCATT |
| R:TTAGAAGACATAAGGAATGA |
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | CF:TCAACTCCATCACATCACAAGAGCTC ATGGAGG- ACC AAGTCAAATC |
| CR:AGCTCCTCGCCCTTGCTCACTCTAGAAAAAAGAG- AA ATTGCAGTAG | |
| erg4B | CF:TCAACTCCATCACATCACAA GAGCTC ATGCCCTC- CA AAAAGGACTC |
| CR:AGCTCCTCGCCCTTGCTCAC TCTAGA GTAAATTC- CA GGAATGATGC | |
| erg4C | NF:GCATGGACGAGCTGTACAAG GAGCTC ATGGATC- GTCCCGGCTTCAT |
| NR:ATGGAGCTATTAAATCACTA TCTAGA TTAGAAG- ACA TACTATAGGA |
Table 2 Primer sequences for GFP fusion vector construction
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | CF:TCAACTCCATCACATCACAAGAGCTC ATGGAGG- ACC AAGTCAAATC |
| CR:AGCTCCTCGCCCTTGCTCACTCTAGAAAAAAGAG- AA ATTGCAGTAG | |
| erg4B | CF:TCAACTCCATCACATCACAA GAGCTC ATGCCCTC- CA AAAAGGACTC |
| CR:AGCTCCTCGCCCTTGCTCAC TCTAGA GTAAATTC- CA GGAATGATGC | |
| erg4C | NF:GCATGGACGAGCTGTACAAG GAGCTC ATGGATC- GTCCCGGCTTCAT |
| NR:ATGGAGCTATTAAATCACTA TCTAGA TTAGAAG- ACA TACTATAGGA |
| Gene | Primer sequence(5' - 3') |
|---|---|
| C-CX | CX1:GGAGA CGTAT TTAGGTGCTA |
| CX2:TGAACTTCA GGGTCAGCTT | |
| N-CX | NX1:CGACAACCAC TACCTGAGCA |
| NX2:TGAAGGGCGT ACTAGGGTTG | |
| erg4A-GFP | F:ATGGAGGACCAAGTCAAATC |
| R:AAAAAGAGAA ATTGCAGTAG | |
| erg4B-GFP | F:ATGCCCTCCAAAAAGGACTC |
| R:GTAAATTCCA GGAATGATGC | |
| erg4C-GFP | F:ATGGATCGTC CCGGCTTCAT |
| R:TTAGAAGACA TAAGGAATGA |
Table 3 Primer sequences for transformant validation
| Gene | Primer sequence(5' - 3') |
|---|---|
| C-CX | CX1:GGAGA CGTAT TTAGGTGCTA |
| CX2:TGAACTTCA GGGTCAGCTT | |
| N-CX | NX1:CGACAACCAC TACCTGAGCA |
| NX2:TGAAGGGCGT ACTAGGGTTG | |
| erg4A-GFP | F:ATGGAGGACCAAGTCAAATC |
| R:AAAAAGAGAA ATTGCAGTAG | |
| erg4B-GFP | F:ATGCCCTCCAAAAAGGACTC |
| R:GTAAATTCCA GGAATGATGC | |
| erg4C-GFP | F:ATGGATCGTC CCGGCTTCAT |
| R:TTAGAAGACA TAAGGAATGA |
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | F:TCCACGAATGTTCGGAATCC |
| R:AGCTCAAACCCAAGAGCATGA | |
| erg4B | F:CGGCGCTTTATGATACCAATG |
| R:GGGAAGGAAGACCTGCAAAAA | |
| erg4C | F:GCATTCCCATCAATCAAAGCA |
| R:CCGGGAAGAAGCACGTAACA | |
| β-tubulin | F:CTCCAGCTCGAGCGTATGAAC |
| R:GGCTCCAAATCGACGAGAAC |
Table 4 Primers for real-time fluorescent quantitative PCR
| Gene | Primer sequence(5' - 3') |
|---|---|
| erg4A | F:TCCACGAATGTTCGGAATCC |
| R:AGCTCAAACCCAAGAGCATGA | |
| erg4B | F:CGGCGCTTTATGATACCAATG |
| R:GGGAAGGAAGACCTGCAAAAA | |
| erg4C | F:GCATTCCCATCAATCAAAGCA |
| R:CCGGGAAGAAGCACGTAACA | |
| β-tubulin | F:CTCCAGCTCGAGCGTATGAAC |
| R:GGCTCCAAATCGACGAGAAC |
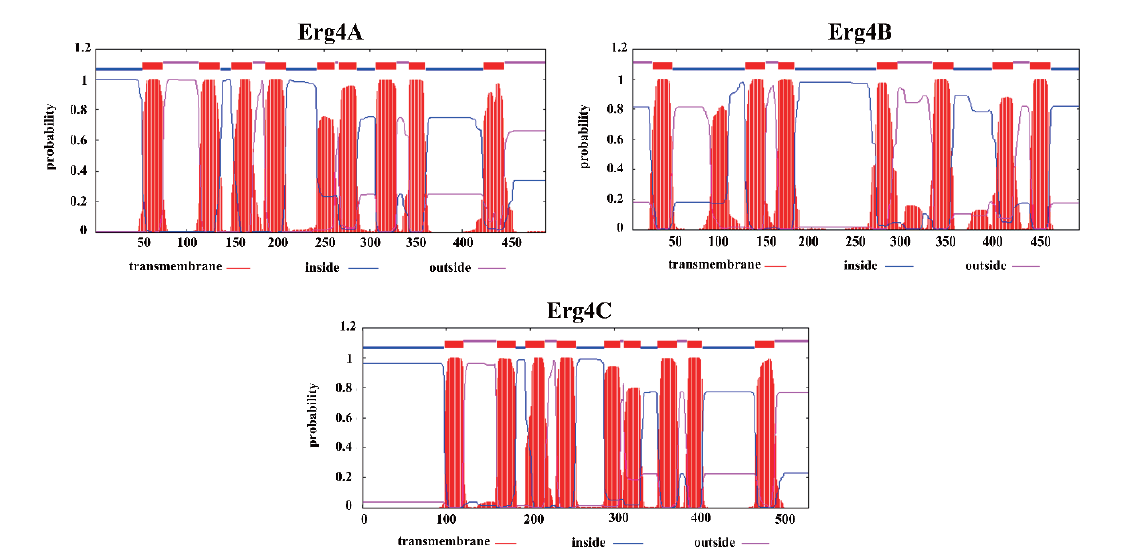
Fig. 3 Transmembrane helices of Erg4A,Erg4B and Erg4C Red rectangle indicates the transmembrane,blue line segment indicates the structure inside the membrane,and the red line segment indicates the structure outside the membrane

Fig. 4 Hydrophilicity/Hydrophobicity of Erg4A,Erg4B and Erg4C protein Positive areas represent the hydrophobic,the negative areas represent the hydrophilic
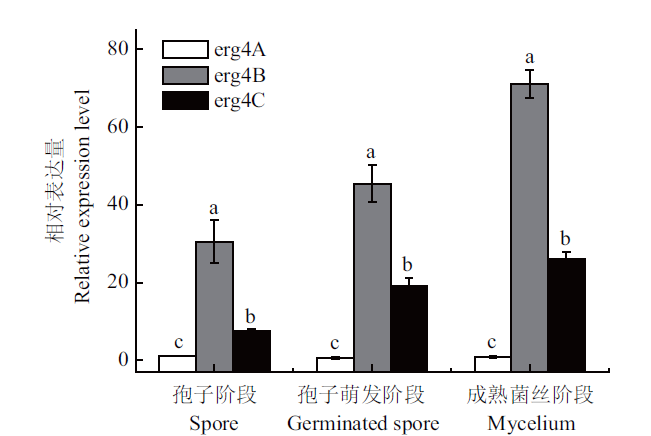
Fig. 10 Relative expressions of erg4A,erg4B and erg4C in the spore,germinated spore and mycelium stages in P. expansum Vertical line indicates standard error(±SE). Different letters refer to significant differences(P<0.05). The same below
| [1] |
Sun C, Fu D, Lu HP, et al. Autoclaved yeast enhances the resistance against Penicillium expansum in postharvest pear fruit and its possible mechanisms of action[J]. Biological Control, 2018, 119:51-58.
doi: 10.1016/j.biocontrol.2018.01.010 URL |
| [2] |
Nierop Groot M, Abee T, van Bokhorst-van de Veen H. Inactivation of conidia from three Penicillium spp. isolated from fruit juices by conventional and alternative mild preservation technologies and disinfection treatments[J]. Food Microbiol, 2019, 81:108-114.
doi: S0740-0020(17)31140-1 pmid: 30910081 |
| [3] |
Hu Z, He B, Ma L, et al. Recent advances in ergosterol biosynjournal and regulation mechanisms in Saccharomyces cerevisiae[J]. Indian J Microbiol, 2017, 57(3):270-277.
doi: 10.1007/s12088-017-0657-1 URL |
| [4] | Sun L, Liao K. The effect of honokiol on ergosterol biosynjournal and vacuole function in Candida albicans[J]. Journal of Microbiol Biotechnology and biotechnology, 2020, 30(12):1835-1842. |
| [5] |
Jin H, McCaffery JM, Grote E. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast[J]. J Cell Biol, 2008, 180(4):813-826.
doi: 10.1083/jcb.200705076 URL |
| [6] | Song J, Zhai P, Zhang Y, et al. The Aspergillus fumigatus damage resistance protein family coordinately regulates ergosterol biosynjournal and azole susceptibility[J]. mBio, 2016, 7(1):e01919-e01915. |
| [7] |
Alcazar-Fuoli L, Mellado E. Ergosterol biosynjournal in Aspergillus fumigatus:its relevance as an antifungal target and role in antifungal drug resistance[J]. Front Microbiol, 2012, 3:439.
doi: 10.3389/fmicb.2012.00439 pmid: 23335918 |
| [8] |
Liu JF, Xia JJ, Nie KL, et al. Outline of the biosynjournal and regulation of ergosterol in yeast[J]. World J Microbiol Biotechnol, 2019, 35(7):98.
doi: 10.1007/s11274-019-2673-2 URL |
| [9] |
Yun Y, Yin D, Dawood DH, et al. Functional characterization of FgERG3 and FgERG5 associated with ergosterol biosynjournal, vegetative differentiation and virulence of Fusarium graminearum[J]. Fungal Genet Biol, 2014, 68:60-70.
doi: 10.1016/j.fgb.2014.04.010 URL |
| [10] |
Jordá T, Puig S. Regulation of ergosterol biosynjournal in Saccharomyces cerevisiae[J]. Genes, 2020, 11(7):795.
doi: 10.3390/genes11070795 URL |
| [11] |
Zweytick D, Hrastnik C, Kohlwein SD, et al. Biochemical characterization and subcellular localization of the sterol C-24(28)reductase, Erg4p, from the yeastSaccharomyces cerevisiae[J]. FEBS Lett, 2000, 470(1):83-87.
pmid: 10722850 |
| [12] |
Feng W, Yang J, Xi Z, et al. Mutations and/or overexpressions of ERG4 and ERG11 genes in clinical azoles-resistant isolates of Candida albicans[J]. Microb Drug Resist, 2017, 23(5):563-570.
doi: 10.1089/mdr.2016.0095 URL |
| [13] |
Tiedje C, Holland DG, Just U, et al. Proteins involved in sterol synjournal interact with Ste20 and regulate cell polarity[J]. J Cell Sci, 2007, 120(pt 20):3613-3624.
doi: 10.1242/jcs.009860 URL |
| [14] |
Aguilar PS, Heiman MG, Walther TC, et al. Structure of sterol aliphatic chains affects yeast cell shape and cell fusion during mating[J]. PNAS, 2010, 107(9):4170-4175.
doi: 10.1073/pnas.0914094107 pmid: 20150508 |
| [15] | Bhattacharya S, Esquivel BD, White TC. Overexpression or deletion of ergosterol biosynjournal genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae[J]. mBio, 2018, 9(4):e01291-e01218. |
| [16] |
Venegas M, Barahona S, González AM, et al. Phenotypic analysis of mutants of ergosterol biosynjournal genes(ERG3 and ERG4)in the red yeast Xanthophyllomyces dendrorhous[J]. Front Microbiol, 2020, 11:1312.
doi: 10.3389/fmicb.2020.01312 URL |
| [17] |
Liu X, Jiang JH, Jiang JH, et al. Sterol C-24(28)reductase, encoded by FgERG4 gene, is important for ergosterol production, hyphal growth, sporulation and virulence in Fusarium graminearum[J]. Molecular plant pathology, 2013, 14(1):71-83.
doi: 10.1111/mpp.2013.14.issue-1 URL |
| [18] | Long NB, Xu XL, Zeng QQ, et al. Erg4A and Erg4B are required for conidiation and azole resistance via regulation of ergosterol biosynjournal in Aspergillus fumigatus[J]. Appl Environ Microbiol, 2017, 83(4):e02924-e02916. |
| [19] |
Li B, Zong Y, Du Z, et al. Genomic characterization reveals insights into patulin biosynjournal and pathogenicity in Penicillium species[J]. Mol Plant Microbe Interact, 2015, 28(6):635-647.
doi: 10.1094/MPMI-12-14-0398-FI URL |
| [20] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4):402-408.
pmid: 11846609 |
| [21] |
Tahir II, Department of Plant Breeding Swedish University of Agricultural Sciences Sweden, Nybom H, et al. Susceptibility to blue mold caused by Penicillium expansum in apple cultivars adapted to a cool climate[J]. Europ J Hort Sci, 2015, 80(3):117-127.
doi: 10.17660/eJHS.2015/80.3.4 URL |
| [22] | 刘馨. 小麦赤霉病菌麦角甾醇生物合成途径中关键基因的功能研究[D]. 杭州:浙江大学, 2012. |
| Liu X. Functional analysis of genes involved in the ergosterol biosynthesis pathway in Fusarium graminearum[D]. Hangzhou:Zhejiang University, 2012. | |
| [23] |
Manchalu S, Mittal N, Spang A, et al. Local translation of yeast ERG4 mRNA at the endoplasmic Reticulum requires the brefeldin A resistance protein Bfr1[J]. RNA, 2019, 25(12):1661-1672.
doi: 10.1261/rna.072017.119 URL |
| [24] | 徐孝苓. 烟曲霉麦角固醇合成相关基因erg4的功能性研究[D]. 南京:南京师范大学, 2017. |
| Xu XL. Functional analysis of ergosterol biosynthesis related gene erg4 in Aspergillus fumigatus[D]. Nanjing:Nanjing Normal University, 2017. | |
| [25] | 于忠洋, 姜洋, 翟明霞, 等. 桑黄麦角甾醇生物合成关键酶PlERG24基因克隆与表达分析[J]. 中草药, 2020, 51(22):5825-5832. |
| Yu ZY, Jiang Y, Zhai MX, et al. Cloning and expression analysis of PlERG24 gene in ergosterol biosynjournal of Phellinus linteus[J]. Chin Tradit Herb Drugs, 2020, 51(22):5825-5832. | |
| [26] |
Rosenfeld E, Beauvoit B. Role of the non-respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae[J]. Yeast, 2003, 20(13):1115-1144.
pmid: 14558145 |
| [27] |
Tang Y, Zhu PK, Lu Z, et al. The photoreceptor components FaWC1 and FaWC2 of fusarium asiaticum cooperatively regulate light responses but play independent roles in virulence expression[J]. Microorganisms, 2020, 8(3):365.
doi: 10.3390/microorganisms8030365 URL |
| [28] |
Casas-Flores S, Rios-Momberg M, Bibbins M, et al. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride[J]. Microbiol Read Engl, 2004, 150(Pt 11):3561-3569.
doi: 10.1099/mic.0.27346-0 URL |
| [1] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [2] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [3] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [4] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [5] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [6] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [7] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| [8] | CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(7): 51-61. |
| [9] | YANG Jia-bao, ZHOU Zhi-ming, ZHANG Zhan, FENG Li, SUN Li. Cloning,Expression of Helianthus annuus HaLACS1 Gene and Identification of Its Functional Complementation in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2022, 38(6): 147-156. |
| [10] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [11] | WANG Nan, ZHANG Rui, PAN Yang-yang, HE Hong-hong, WANG Jing-lei, CUI Yan, YU Si-jiu. Cloning of Bos grunniens TGF-β1 Gene and Its Expression in Major Organs of Female Reproductive System [J]. Biotechnology Bulletin, 2022, 38(6): 279-290. |
| [12] | LI Yu-hang, WANG Xing-ping, YANG Jian, LUORENG Zhuo-ma, REN Qian-qian, WEI Da-wei, MA Yun. Expression and Functional Analysis of miR-665 in Bovine Mammary Epithelial Cell Inflammation [J]. Biotechnology Bulletin, 2022, 38(5): 159-168. |
| [13] | LI Yang, ZHANG Xiao-tian, PIAO Jing-zi, ZHOU Ru-jun, LI Zi-bo, GUAN Hai-wen. Cloning and Bioinformatics Analysis of Blue-light Receptor EaWC 1 Gene in Elsinoë arachidis [J]. Biotechnology Bulletin, 2022, 38(5): 93-99. |
| [14] | HAO Qing-qing, YAO Sheng, LIU Jia-he, CHEN Pei-zhen, ZHANG Meng-yang, JI Kong-shu. Cloning and Expression Analysis of NAC Transcription Factor PmNAC8 in Pinus massoniana [J]. Biotechnology Bulletin, 2022, 38(4): 202-216. |
| [15] | ZHANG Lin, WEI Zhen-zhen, SONG Cheng-wei, GUO Li-li, GUO Qi, HOU Xiao-gai, WANG Hua-fang. Cloning and Expression Analysis of PoFD Gene from Paeonia ostii ‘Fengdan’ [J]. Biotechnology Bulletin, 2022, 38(11): 104-111. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||