








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (1): 262-269.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0484
Previous Articles Next Articles
XIE Hong1( ), ZHOU Li-ying2, LI Shu-wen1, WANG Meng-di1, AI Ye1, CHAO Yue-hui1(
), ZHOU Li-ying2, LI Shu-wen1, WANG Meng-di1, AI Ye1, CHAO Yue-hui1( )
)
Received:2023-05-23
Online:2024-01-26
Published:2024-02-06
Contact:
CHAO Yue-hui
E-mail:xiehong20211014@163.com;chaoyuehui@bjfu.edu.cn
XIE Hong, ZHOU Li-ying, LI Shu-wen, WANG Meng-di, AI Ye, CHAO Yue-hui. Structural and Functional Analysis of MtCIM Gene in Medicago truncatula[J]. Biotechnology Bulletin, 2024, 40(1): 262-269.
| 引物名称 Prime name | 引物序列 Prime sequence(5'-3') |
|---|---|
| T7 | TAATACGACTCACTATAGG |
| 3-BD | TAAGAGTCACTTTAAAATTTGTAT |
| pGBKT7-CIM-F | GCTATTTCTACTGATTTTTCATGGCTCTTACACTTGAACA |
| pGBKT7-CIM-R | CAAAATCATGTCAAGGTCTTTTAAAAATTCACAATTGACC |
| 3302Y-F | TGACGCACAATCCCACTATCCTT |
| 3302Y-R | CCGTCCAGCTCGACCAGGAT |
| 3302Y-MtCIM-F | GAACACGGGGGACTCTTGACCATGGCTCTTACACTTGAACA |
| 3302Y-MtCIM-R | ACGCGTACTAGTCAGATCTACAAAATTCACAATTGACCTAT |
| MtActin-F | TGATCTGGCTGGTCGTGACCTTA |
| MtActin-R | ATGCCTGCTGCTTCCATTCCTAT |
| MtCIM-RT-F | TGGTGACGGAAGCGAAGGTGGTG |
| MtCIM-RT-R | CTTAACAGGGTTGCCTGAACATA |
Table 1 Primer sequences
| 引物名称 Prime name | 引物序列 Prime sequence(5'-3') |
|---|---|
| T7 | TAATACGACTCACTATAGG |
| 3-BD | TAAGAGTCACTTTAAAATTTGTAT |
| pGBKT7-CIM-F | GCTATTTCTACTGATTTTTCATGGCTCTTACACTTGAACA |
| pGBKT7-CIM-R | CAAAATCATGTCAAGGTCTTTTAAAAATTCACAATTGACC |
| 3302Y-F | TGACGCACAATCCCACTATCCTT |
| 3302Y-R | CCGTCCAGCTCGACCAGGAT |
| 3302Y-MtCIM-F | GAACACGGGGGACTCTTGACCATGGCTCTTACACTTGAACA |
| 3302Y-MtCIM-R | ACGCGTACTAGTCAGATCTACAAAATTCACAATTGACCTAT |
| MtActin-F | TGATCTGGCTGGTCGTGACCTTA |
| MtActin-R | ATGCCTGCTGCTTCCATTCCTAT |
| MtCIM-RT-F | TGGTGACGGAAGCGAAGGTGGTG |
| MtCIM-RT-R | CTTAACAGGGTTGCCTGAACATA |
| 顺式元件Cis-element | 基序Motif(5'-3') | 位置Position | 功能Function |
|---|---|---|---|
| ABRE | CACGTG | 348 | 脱落酸响应元件ABA response element |
| ACGTC | 349 | ||
| ACGTC | 708 | ||
| P-box | CCTTTTG | 53 | 赤霉素响应元件Gibberellin response element |
| TC-rich repeats | GTTTTCTTAC | 103 | 防御和胁迫反应相关的元件Defense and stress response related elements |
| TGA-element | AACGAC | 510 | 生长素响应元件Auxin response element |
| TCT-motif | TCTTAC | 103 | 光响应元件Light response element |
| ARE | AAACCA | 567 | 厌氧诱导元件Anaerobic induction element |
| TATA-box | TATTTAAA | 21 | 转录起始点-30附近的核心启动子元件 Core promoter element near trabscription starting point -30 |
| TATAA | 148 | ||
| TATTA | 633 | ||
| TATA | 502 | ||
| TATATA | 87 | ||
| TATA | 275 | ||
| G-BOX | CACGTG | 348 | 参与光反应的顺式作用元件Cis-acting elements involved in photoreactions |
| CACGTG | 707 |
Table 2 The main cis-acting elements of MtCIM promoter
| 顺式元件Cis-element | 基序Motif(5'-3') | 位置Position | 功能Function |
|---|---|---|---|
| ABRE | CACGTG | 348 | 脱落酸响应元件ABA response element |
| ACGTC | 349 | ||
| ACGTC | 708 | ||
| P-box | CCTTTTG | 53 | 赤霉素响应元件Gibberellin response element |
| TC-rich repeats | GTTTTCTTAC | 103 | 防御和胁迫反应相关的元件Defense and stress response related elements |
| TGA-element | AACGAC | 510 | 生长素响应元件Auxin response element |
| TCT-motif | TCTTAC | 103 | 光响应元件Light response element |
| ARE | AAACCA | 567 | 厌氧诱导元件Anaerobic induction element |
| TATA-box | TATTTAAA | 21 | 转录起始点-30附近的核心启动子元件 Core promoter element near trabscription starting point -30 |
| TATAA | 148 | ||
| TATTA | 633 | ||
| TATA | 502 | ||
| TATATA | 87 | ||
| TATA | 275 | ||
| G-BOX | CACGTG | 348 | 参与光反应的顺式作用元件Cis-acting elements involved in photoreactions |
| CACGTG | 707 |
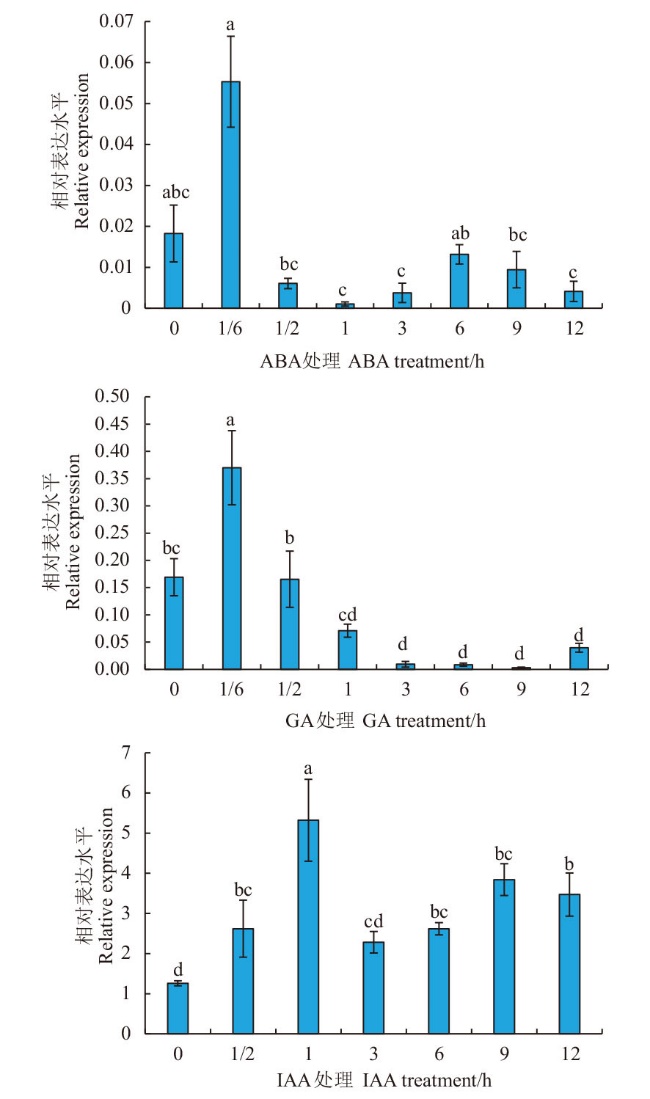
Fig. 4 MtCIM expression analysis in Medicago truncatula under different hormone treatments Different lowercase letters indicate a significant difference at the 0.05 level

Fig. 7 Difference between transgenic and wild type plants under total phosphorus and low phosphorus treatment CIM: Transgenic plants. WT: Wild types. A, C: Total phosphorus. B, D: Low phosphorus
| [1] |
McQueen-Mason S, Cosgrove DJ. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension[J]. PNAS, 1994. 91(14): 6574-6578.
doi: 10.1073/pnas.91.14.6574 pmid: 11607483 |
| [2] | 雷忠萍. 棉花扩展蛋白基因的鉴定及特征研究[D]. 杨凌: 西北农林科技大学, 2018. |
| Lei ZP. Identification and characterization of cotton expansin gene[D]. Yangling: Northwest A&F University, 2018. | |
| [3] |
Lee Y, Choi D, Kende H. Expansins: ever-expanding numbers and functions[J]. Curr Opin Plant Biol, 2001, 4(6): 527-532.
doi: 10.1016/s1369-5266(00)00211-9 pmid: 11641069 |
| [4] |
Yan A, Wu MJ, Yan LM. AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis[J]. PLoS One, 2014, 9(1): e85208.
doi: 10.1371/journal.pone.0085208 URL |
| [5] |
Cho HT, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana[J]. PNAS, 2000, 97(17): 9783-9788.
doi: 10.1073/pnas.160276997 pmid: 10931949 |
| [6] |
Lee HW, Kim MJ, Kim NY, et al. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis[J]. Plant J, 2013. 73(2): 212-224.
doi: 10.1111/tpj.2013.73.issue-2 URL |
| [7] |
Lee HW, Kim J. EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response[J]. Plant Cell Physiol, 2013, 54(10): 1600-1611.
doi: 10.1093/pcp/pct105 pmid: 23872272 |
| [8] |
Ma N, Wang Y, Qiu SC, et al. Overexpression of OsEXPA8, a root-specific gene, improves rice growth and root system architecture by facilitating cell extension[J]. PloS one, 2013, 8(10): e75997.
doi: 10.1371/journal.pone.0075997 URL |
| [9] |
Chen YH, Han YY, Zhang M, et al. Overexpression of the wheat expansin gene TaEXPA2 improved seed production and drought tolerance in transgenic tobacco plants[J]. PLos One, 2016, 11(4): e0153494.
doi: 10.1371/journal.pone.0153494 URL |
| [10] |
Kong YB, Wang B, Du H, et al. GmEXLB1, a soybean expansin-like B gene, alters root architecture to improve phosphorus acquisition in Arabidopsis[J]. Front Plant Sci, 2019, 10: 808.
doi: 10.3389/fpls.2019.00808 URL |
| [11] |
Yang ZJ, Gao Z, Zhou HW, et al. GmPTF1 modifies root architecture responses to phosphate starvation primarily through regulating GmEXPB2 expression in soybean[J]. Plant J, 2021, 107(2): 525-543.
doi: 10.1111/tpj.v107.2 URL |
| [12] |
Sampedro J, Cosgrove DJ. The expansin superfamily[J]. Genome Biol, 2005, 6(12): 242.
doi: 10.1186/gb-2005-6-12-242 pmid: 16356276 |
| [13] | 杨君娇. 扩展蛋白基因TaEXPA2在小麦耐旱性中的功能分析[D]. 泰安: 山东农业大学, 2020. |
| Yang JJ. Functional analysis of expandable protein gene TaEXPA2 in drought tolerance of wheat[D]. Tai'an: Shandong Agricultural University, 2020. | |
| [14] | Chen LJ, Zou WS, Fei CY, et al. α-expansin EXPA4 positively regulates abiotic stress tolerance but negatively regulates pathogen resistance in Nicotiana tabacum[J]. Plant Cell Physiol, 2018, 59(11): 2317-2330. |
| [15] |
Zhang BY, Chang L, Sun WN, et al. Overexpression of an expansin-like gene, GhEXLB2 enhanced drought tolerance in cotton[J]. Plant Physiol Biochem, 2021, 162: 468-475.
doi: 10.1016/j.plaphy.2021.03.018 URL |
| [16] |
Winey M, Meehl JB, O'Toole ET, et al. Conventional transmission electron microscopy[J]. Mol Biol Cell, 2014, 25(3): 319-323.
doi: 10.1091/mbc.E12-12-0863 pmid: 24482357 |
| [17] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [18] | 艾晔, 钱旭, 韩烈保, 等. 蒺藜苜蓿CIM基因的克隆及转化[J/OL]. 分子植物育种, 2021. https://kns.cnki.net/kcms/detail/46.1068.S.20210630.0948.004.html. |
| Ai Y, Qian X, Han LB, et al. Cloning and transformation of CIM gene from Medicago truncatula[J/OL]. Mol Plant Breed, 2021. https://kns.cnki.net/kcms/detail/46.1068.S.20210630.0948.004.html. | |
| [19] | 郑珊珊, 石卓兴, 代琦, 等. 蛋白质亚细胞定位预测研究进展[J]. 科技视界, 2014(15): 183-184. |
| Zheng SS, Shi ZX, Dai Q, et al. Research progress of subcellular localization prediction in protein[J]. Sci Technol Vis, 2014(15): 183-184. | |
| [20] | 李杰, 张福城, 王文泉, 等. 高等植物启动子的研究进展[J]. 生物技术通讯, 2006, 17(4): 658-661. |
| Li J, Zhang FC, Wang WQ, et al. Advance in the study of higher plant promoter[J]. Lett Biotechnol, 2006, 17(4): 658-661. | |
| [21] | 张春晓, 王文棋, 蒋湘宁, 等. 植物基因启动子研究进展[J]. 遗传学报, 2004, 31(12): 1455-1464. |
| Zhang CX, Wang WQ, Jiang XN, et al. Review on plant gene promoters[J]. Acta Genet Sin, 2004, 31(12): 1455-1464. | |
| [22] |
冯寒骞, 李超. 生长素信号转导研究进展[J]. 生物技术通报, 2018, 34(7): 24-30.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-0488 |
| Feng HL, Li C. Research advances of auxin signal transduction[J]. 2018, 34(7): 24-30. | |
| [23] | 韩阳阳, 王天晓, 王玮. 扩展蛋白与激素调控的生长发育[J]. 生命的化学, 2015. 35(6): 778-784. |
| Han YY, Wang TX, Wang W. Research progress on the relationship between expansins and phytohormone induced plant growth and development[J]. Chemistry life, 2015, 35(6): 778-784. | |
| [24] | 孟永娇, 李季, 徐建, 等, 黄瓜扩张素蛋白基因CsEXPb1的克隆及表达分析[J]. 园艺学报, 2015, 42(4): 679-688. |
| Meng YJ, Li J, Xu J, et al. Cloning and expression analysis of the expansin gene CsEXPb1 in Cucumber[J]. Acta Hortic Sin, 2015, 42(4): 679-688. | |
| [25] |
Wu Y, Meeley RB, Cosgrove DJ. Analysis and expression of theα-expansin and β-expansin gene families in maize[J]. Plant Physiol, 2001, 126(1): 222-232.
pmid: 11351085 |
| [26] | 丁安明, 陈志华, 杨懿德, 等. NtabEXPA12基因过表达对烟草叶片发育及抗逆性的影响[J]. 中国烟草科学, 2021, 42(4): 58-66. |
| Ding AM, Chen ZH, Yang YD, et al. Overexpression of Ntab EXPA12 affects leaf development and abiotic stress tolerance in tobacco[J]. Chin tob sci, 2021, 42(4): 58-66. | |
| [27] |
Tan J, Wang ML, Shi ZY, et al. OsEXPA10 mediates the balance between growth and resistance to biotic stress in rice[J]. Plant Cell Rep, 2018, 37(7): 993-1002.
doi: 10.1007/s00299-018-2284-7 pmid: 29619515 |
| [28] |
Cheung AY, Cosgrove DJ, Hara-Nishimura I, et al. A rich and bountiful harvest: Key discoveries in plant cell biology[J]. Plant Cell, 2022, 34(1): 53-71.
doi: 10.1093/plcell/koab234 URL |
| [29] | 邢国芳, 郭平毅. 植物响应低磷胁迫的功能基因组研究进展[J]. 生物技术通报, 2013(7): 1-6. |
| Xing GF, Guo PY. Functional genomics approaches for the study of low phosphate responses in higher plants[J]. Biotechnol Bull, 2013(7): 1-6. |
| [1] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [2] | TANG Wei-lin, KANG Qin, WANG Xia, SHEN Ming-yang, SUN Xin-jiang, WANG Ke, HOU Kai, WU Wei, XU Dong-bei. Cloning and Expression Pattern Analysis of Jasmonic Acid Receptor Gene McCOI1a in Mentha canadensis L. [J]. Biotechnology Bulletin, 2024, 40(1): 270-280. |
| [3] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [4] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [5] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [6] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [7] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [8] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [9] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [10] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [11] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [12] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [13] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [14] | TENG Meng-xin, XU Ya, HE Jing, WANG Qi, QIAO Fei, LI Jing-yang, LI Xin-guo. Cloning and Prokaryotic Expression Analysis of MaMC6 in Banana [J]. Biotechnology Bulletin, 2023, 39(12): 179-186. |
| [15] | SHANG Yi-tong, YAN Huan-huan, WANG Li-hong, TIAN Xue-qin, XUE Ping-hong, LUO Tao, HU Zhi-hong. Study on the Function of Phosphomevalonate Kinase in Aspergillus oryzae [J]. Biotechnology Bulletin, 2023, 39(12): 311-319. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||