








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (1): 270-280.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0789
Previous Articles Next Articles
TANG Wei-lin1( ), KANG Qin1, WANG Xia2, SHEN Ming-yang1, SUN Xin-jiang1, WANG Ke1, HOU Kai1, WU Wei1, XU Dong-bei1(
), KANG Qin1, WANG Xia2, SHEN Ming-yang1, SUN Xin-jiang1, WANG Ke1, HOU Kai1, WU Wei1, XU Dong-bei1( )
)
Received:2023-08-14
Online:2024-01-26
Published:2024-02-06
Contact:
XU Dong-bei
E-mail:weilintang2021@163.com;xudongbei2006@126.com
TANG Wei-lin, KANG Qin, WANG Xia, SHEN Ming-yang, SUN Xin-jiang, WANG Ke, HOU Kai, WU Wei, XU Dong-bei. Cloning and Expression Pattern Analysis of Jasmonic Acid Receptor Gene McCOI1a in Mentha canadensis L.[J]. Biotechnology Bulletin, 2024, 40(1): 270-280.
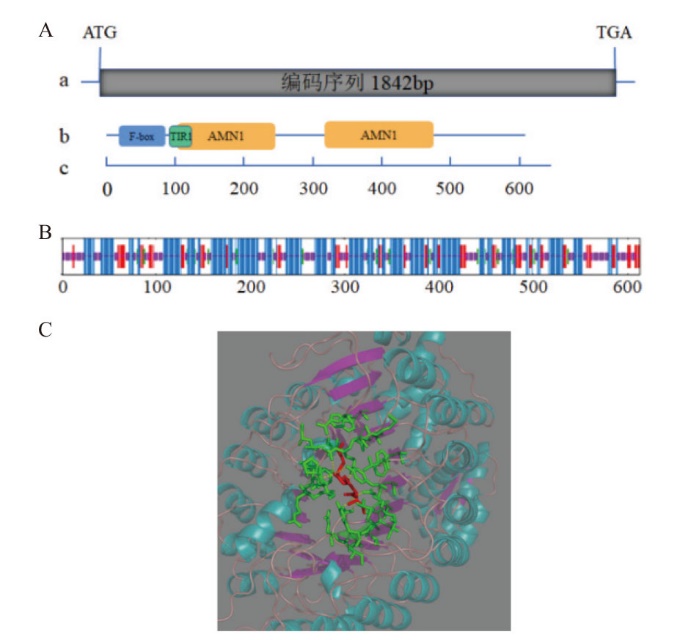
Fig. 1 McCOI1a gene and its protein structure analysis in M. canadensis A: The diagram of McCOI1a gene and the conserved domain of McCOI1a protein. a: The full-length of McCOI1a; b: the full-length sequence of protein and the positions of conserved domain F-box, TIR1, and AMN1; c: the scale bar of the length of amino acid; bp: base pair. B: Secondary structure of McCOI1a protein. Blue: α-helix; Green: beta-turn; Red: extended-strand; Purple: random coil. C: Tertiary structure of McCOI1a protein. Baby blue: α-helix. Purple: beta-turn; Green: the residue around the ligand; Red: ligand; The dotted yellow line: hydrogen bond
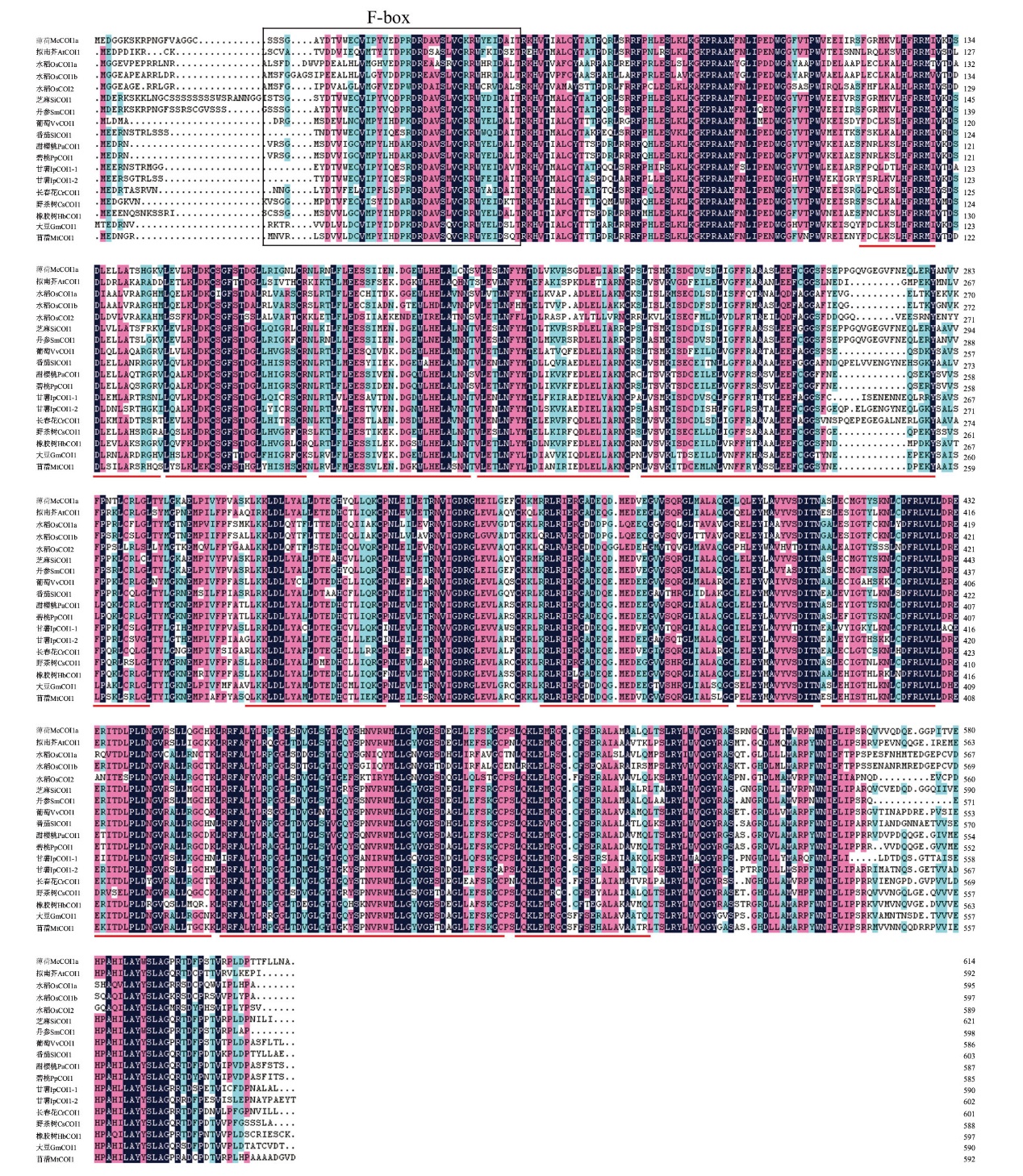
Fig. 2 Amino acid sequence comparison of McCOI1a and its homologous proteins Black box: F-box domain; red underline: leucine-rich repeat sequence(LRR)

Fig. 6 Expression analysis of McCOI1a gene in different tissues and phyllotaxis of M. canadensis A: Expressions of McCOI1a in the roots, stems, young leaf, old leaf, flower, bud, apical shoot of M. canadensis. B: Relative expressions of McCOI1a in the leaves at different positions. Leaf 1-leaf 8: The first to eighth leaf morphologically from top to bottom. The data are of ±SE of three biological replicates

Fig. 7 Expression analysis of McCOI1a in the leaves of M. canadensis under MeJA and abiotic stress treatments The data ±SE(n=3)of three biological replicates. Duncan test was used in ANOVA, the lowercase letters indicate significant differences at the 0.05 level. The same below
| [1] | 陈智坤, 梁呈元, 任冰如, 等. 薄荷属植物挥发性成分及药理作用研究进展[J]. 天然产物研究与开发, 2013, 25(6): 856-861, 865. |
| Chen ZK, Liang CY, Ren BR, et al. Advances in the studies of chemical constituents and pharmacological activities of volatile components from Mentha L[J]. Nat Prod Res Dev, 2013, 25(6): 856-861, 865. | |
| [2] |
Yu X, Liang CY, Chen J, et al. The effects of salinity stress on morphological characteristics, mineral nutrient accumulation and essential oil yield and composition in Mentha canadensis L[J]. Sci Hortic, 2015, 197: 579-583.
doi: 10.1016/j.scienta.2015.10.023 URL |
| [3] |
Azimychetabi Z, Sabokdast Nodehi M, Karami Moghadam T, et al. Cadmium stress alters the essential oil composition and the expression of genes involved in their synthesis in peppermint(Mentha piperita L.)[J]. Ind Crops Prod, 2021, 168: 113602.
doi: 10.1016/j.indcrop.2021.113602 URL |
| [4] |
Singh R, Luxmi S, Charak A, et al. Effects of intermittent drought on the essential oil yield, contents, and nutrient status of Mentha longifolia(L.) huds[J]. J Essent Oil Bear Plants, 2022, 25(3): 626-638.
doi: 10.1080/0972060X.2022.2091957 URL |
| [5] |
Yu XX, Zhang WJ, Zhang Y, et al. The roles of methyl jasmonate to stress in plants[J]. Funct Plant Biol, 2019, 46(3): 197-212.
doi: 10.1071/FP18106 pmid: 32172764 |
| [6] |
Yan JB, Zhang C, Gu M, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor[J]. Plant Cell, 2009, 21(8): 2220-2236.
doi: 10.1105/tpc.109.065730 URL |
| [7] |
Wasternack C, Song SS. Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription[J]. J Exp Bot, 2017, 68(6): 1303-1321.
doi: 10.1093/jxb/erw443 pmid: 27940470 |
| [8] |
Xie DX, Feys BF, James S, et al. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility[J]. Science, 1998, 280(5366): 1091-1094.
doi: 10.1126/science.280.5366.1091 pmid: 9582125 |
| [9] |
Ye M, Luo SM, Xie JF, et al. Silencing COI1 in rice increases susceptibility to chewing insects and impairs inducible defense[J]. PLoS One, 2012, 7(4): e36214.
doi: 10.1371/journal.pone.0036214 URL |
| [10] |
Lee SH, Sakuraba Y, Lee T, et al. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b(OsCOI1b)delays leaf senescence[J]. J Integr Plant Biol, 2015, 57(6): 562-576.
doi: 10.1111/jipb.v57.6 URL |
| [11] |
Bai JF, Wang YK, Wang P, et al. Genome-wide identification and analysis of the COI gene family in wheat(Triticum aestivum L.)[J]. BMC Genomics, 2018, 19(1): 754.
doi: 10.1186/s12864-018-5116-9 |
| [12] |
Qi XL, Guo SW, Wang D, et al. ZmCOI2a and ZmCOI2b redundantly regulate anther dehiscence and gametophytic male fertility in maize[J]. Plant J, 2022, 110(3): 849-862.
doi: 10.1111/tpj.v110.3 URL |
| [13] |
Sun TT, Meng YT, Cen GL, et al. Genome-wide identification and expression analysis of the coronatine-insensitive 1(COI1)gene family in response to biotic and abiotic stresses in Saccharum[J]. BMC Genomics, 2022, 23(1): 38.
doi: 10.1186/s12864-021-08255-0 |
| [14] |
Chen QF, Dai LY, Xiao S, et al. The COI1 and DFR genes are essential for regulation of jasmonate-induced anthocyanin accumulation in Arabidopsis[J]. J Integr Plant Biol, 2007, 49(9): 1370-1377.
doi: 10.1111/jipb.2007.49.issue-9 URL |
| [15] |
Shen Q, Lu X, Yan TX, et al. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua[J]. New Phytol, 2016, 210(4): 1269-1281.
doi: 10.1111/nph.2016.210.issue-4 URL |
| [16] |
Zhou YY, Sun W, Chen JF, et al. SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza[J]. Sci Rep, 2016, 6: 22852.
doi: 10.1038/srep22852 |
| [17] |
Zhang HT, Hedhili S, Montiel G, et al. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus[J]. Plant J, 2011, 67(1): 61-71.
doi: 10.1111/tpj.2011.67.issue-1 URL |
| [18] |
Zhang HB, Bokowiec MT, Rushton PJ, et al. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis[J]. Mol Plant, 2012, 5(1): 73-84.
doi: 10.1093/mp/ssr056 URL |
| [19] |
Xu DB, Ma YN, Qin TF, et al. Transcriptome-wide identification and characterization of the JAZ gene family in Mentha canadensis L[J]. Int J Mol Sci, 2021, 22(16): 8859.
doi: 10.3390/ijms22168859 URL |
| [20] |
Ma YN, Xu DB, Li L, et al. Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua[J]. Sci Adv, 2018, 4(11): eaas9357.
doi: 10.1126/sciadv.aas9357 URL |
| [21] |
Ruan JJ, Zhou YX, Zhou ML, et al. Jasmonic acid signaling pathway in plants[J]. Int J Mol Sci, 2019, 20(10): 2479.
doi: 10.3390/ijms20102479 URL |
| [22] |
Chini A, Boter M, Solano R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module[J]. FEBS J, 2009, 276(17): 4682-4692.
doi: 10.1111/j.1742-4658.2009.07194.x pmid: 19663905 |
| [23] |
Trang Nguyen H, Thi Mai To H, Lebrun M, et al. Jasmonates-the master regulator of rice development, adaptation and defense[J]. Plants, 2019, 8(9): 339.
doi: 10.3390/plants8090339 URL |
| [24] |
Chen J, Yang HT, Ma S, et al. HbCOI1 perceives jasmonate to trigger signal transduction in Hevea brasiliensis[J]. Tree Physiol, 2021, 41(3): 460-471.
doi: 10.1093/treephys/tpaa124 URL |
| [25] |
Hu S, Yu KM, Yan JB, et al. Jasmonate perception: ligand-receptor interaction, regulation, and evolution[J]. Mol Plant, 2023, 16(1): 23-42.
doi: 10.1016/j.molp.2022.08.011 URL |
| [26] | 付鸿博, 向成刚, 陶宏征, 等. 灯盏花COI1基因的克隆及表达分析[J]. 分子植物育种, 2023. http://kns.cnki.net/kcms/detail/46.1068.s.20220707.1441.006.html. |
| Fu HB, Xiang CG, Tao HZ, et al. Cloning and expression analysis of COI1 gene in Erigeron breviscapus[J]. Molecular Plant Breeding, 2023. http://kns.cnki.net/kcms/detail/46.1068.s.20220707.1441.006.html. | |
| [27] |
Lee HY, Seo JS, Cho JH, et al. Oryza sativa COI homologues restore jasmonate signal transduction in Arabidopsis coi1-1 mutants[J]. PLoS One, 2013, 8(1): e52802.
doi: 10.1371/journal.pone.0052802 URL |
| [28] | 吴晨雨, 何俊娜, 钟雄辉, 等. 唐菖蒲茉莉素受体基因GhCOI1的克隆与表达分析[J]. 园艺学报, 2015, 42(2): 289-300. |
| Wu CY, He JN, Zhong XH, et al. Cloning and expression analysis of coronatine insensitive gene encoding a jasmonate receptor from Gladiolus hybridus[J]. Acta Hortic Sin, 2015, 42(2): 289-300. | |
| [29] |
Shan XY, Wang JX, Chua L, et al. The role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence[J]. Plant Physiol, 2011, 155(2): 751-764.
doi: 10.1104/pp.110.166595 URL |
| [30] |
Chen YN, Feng PP, Tang BY, et al. The AP2/ERF transcription factor SlERF.F5 functions in leaf senescence in tomato[J]. Plant Cell Rep, 2022, 41(5): 1181-1195.
doi: 10.1007/s00299-022-02846-1 pmid: 35238951 |
| [31] |
Liao YC, Wei JH, Xu YH, et al. Cloning, expression and characterization of COI1 gene(AsCOI1)from Aquilaria sinensis(Lour.) Gilg[J]. Acta Pharm Sin B, 2015, 5(5): 473-481.
doi: 10.1016/j.apsb.2015.05.009 URL |
| [32] |
Liu R, Wang JB, Xiao M, et al. AaCOI1, encoding a CORONATINE INSENSITIVE 1-like protein of Artemisia annua L., is involved in development, defense, and anthocyanin synthesis[J]. Genes, 2020, 11(2): 221.
doi: 10.3390/genes11020221 URL |
| [33] |
Yastreb TO, Kolupaev YE, Shkliarevskyi MA, et al. Involvement of jasmonate signaling components in salt stress-induced stomatal closure in Arabidopsis thaliana[J]. Cytol Genet, 2020, 54(4): 318-323.
doi: 10.3103/S009545272004012X |
| [34] |
Gupta A, Bhardwaj M, Tran LS P. JASMONATE ZIM-DOMAIN family proteins: important nodes in jasmonic acid-abscisic acid crosstalk for regulating plant response to drought[J]. Curr Protein Pept Sci, 2021, 22(11): 759-766.
doi: 10.2174/1389203722666211018114443 URL |
| [35] |
Chen YJ, Chen Y, Shi ZJ, et al. Biosynthesis and signal transduction of ABA, JA, and BRs in response to drought stress of Kentucky bluegrass[J]. Int J Mol Sci, 2019, 20(6): 1289.
doi: 10.3390/ijms20061289 URL |
| [36] |
Valenzuela CE, Acevedo-Acevedo O, Miranda GS, et al. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root[J]. J Exp Bot, 2016, 67(14): 4209-4220.
doi: 10.1093/jxb/erw202 pmid: 27217545 |
| [37] |
Yang ZB, He CM, Ma YQ, et al. Jasmonic acid enhances Al-induced root growth inhibition[J]. Plant Physiol, 2017, 173(2): 1420-1433.
doi: 10.1104/pp.16.01756 URL |
| [38] |
Zhang CH, Huang RQ, Zhan NH, et al. Methyl jasmonate and selenium synergistically mitigative cadmium toxicity in hot pepper(Capsicum annuum L.) plants by improving antioxidase activities and reducing Cd accumulation[J]. Environ Sci Pollut Res Int, 2023, 30(34): 82458-82469.
doi: 10.1007/s11356-023-28273-7 |
| [39] |
Hanaka A, Wójcik M, Dresler S, et al. Does methyl jasmonate modify the oxidative stress response in Phaseolus coccineus treated with Cu?[J]. Ecotoxicol Environ Saf, 2016, 124: 480-488.
doi: 10.1016/j.ecoenv.2015.11.024 URL |
| [1] | XIE Hong, ZHOU Li-ying, LI Shu-wen, WANG Meng-di, AI Ye, CHAO Yue-hui. Structural and Functional Analysis of MtCIM Gene in Medicago truncatula [J]. Biotechnology Bulletin, 2024, 40(1): 262-269. |
| [2] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [3] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [4] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [5] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [6] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [7] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [8] | WEI Ming WANG Xin-yu WU Guo-qiang ZHAO Meng. The Role of NAD-dependent Deacetylase SRT in Plant Epigenetic Inheritance Regulation [J]. Biotechnology Bulletin, 2023, 39(4): 59-70. |
| [9] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [10] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [11] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [12] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [13] | YAN Xiong-ying, WANG Zhen, WANG Xia, YANG Shi-hui. Microbial Sulfur Metabolism and Stress Resistance [J]. Biotechnology Bulletin, 2023, 39(11): 150-167. |
| [14] | ZHANG Hong-hong, FANG Xiao-feng. Advances in the Regulation of Stress Sensing and Responses by Phase Separation in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 44-53. |
| [15] | GE Wen-dong, WANG Teng-hui, MA Tian-yi, FAN Zhen-yu, WANG Yu-shu. Genome-wide Identification of the PRX Gene Family in Cabbage(Brassica oleracea L. var. capitata)and Expression Analysis Under Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 252-260. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||