








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (2): 1-8.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0963
Received:2023-10-18
Online:2024-02-26
Published:2024-03-13
CHEN Yan-mei. Crosstalk Between Different Post-translational Modifications and Its Regulatory Mechanisms in Plant Growth and Development[J]. Biotechnology Bulletin, 2024, 40(2): 1-8.
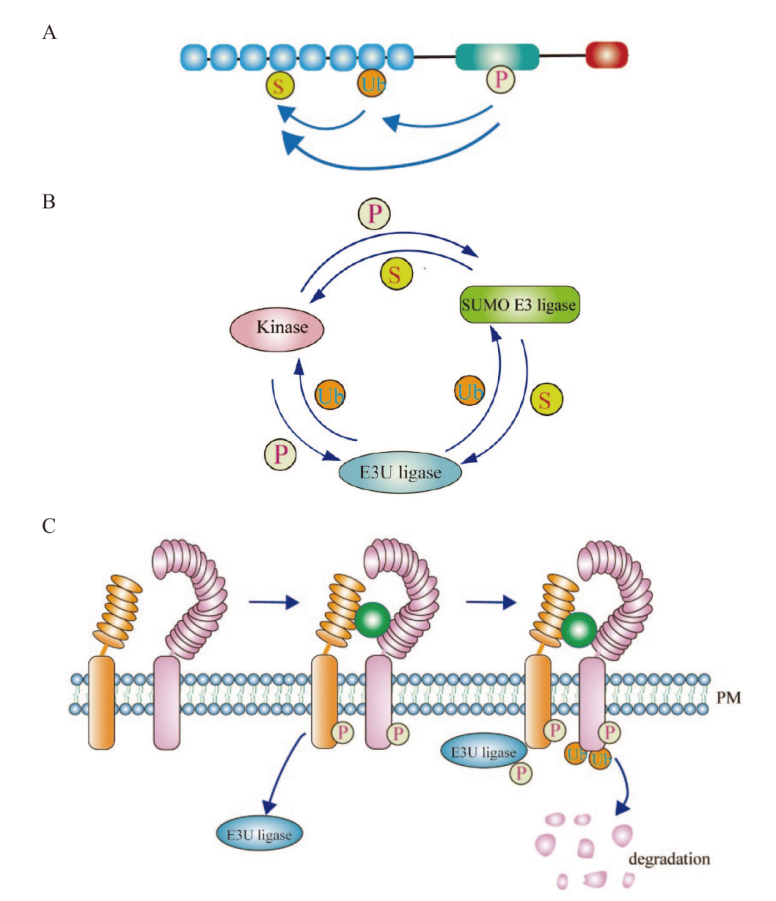
Fig. 1 Crosstalk and interaction among “Phosphorylation-ubiquitination-sumoylation” A: “Phosphorylation-ubiquitination-sumoylation” function at different amino acid residues of the same protein. The molecular function of different modifications among proteins is jointly regulated through mutual antagonism or promotion. B: “Phosphorylation-ubiquitination-sumoylation” are the modifications conducted through the use of different proteins or enzymes as substrates. C: The signaling processes of receptor-like kinase are co-regulated by “phosphorylation-ubiquitination”. P: Phosphorylation; S: sumoylation; Ub: ubiquitination; PM: plasma membrane
| [1] |
Chen YM, Wang Y, Yang J, et al. Exploring the diversity of plant proteome[J]. J Integr Plant Biol, 2021, 63(7): 1197-1210.
doi: 10.1111/jipb.13087 |
| [2] |
Chen YM, Hoehenwarter W. Rapid and reproducible phosphopeptide enrichment by tandem metal oxide affinity chromatography: application to boron deficiency induced phosphoproteomics[J]. Plant J, 2019, 98(2): 370-384.
doi: 10.1111/tpj.2019.98.issue-2 URL |
| [3] |
Consortium TU. UniProt: the universal protein knowledgebase[J]. Nucleic Acids Res, 2017, 45(D1): D158-D169.
doi: 10.1093/nar/gkw1099 URL |
| [4] |
Willems P, Horne A, Van Parys T, et al. The Plant PTM Viewer, a central resource for exploring plant protein modifications[J]. Plant J, 2019, 99(4): 752-762.
doi: 10.1111/tpj.v99.4 URL |
| [5] |
Millar AH, Heazlewood JL, Giglione C, et al. The scope, functions, and dynamics of posttranslational protein modifications[J]. Annu Rev Plant Biol, 2019, 70: 119-151.
doi: 10.1146/annurev-arplant-050718-100211 pmid: 30786234 |
| [6] |
Chen YM, Wang Y. Mapping histone modification-dependent protein interactions with chemical proteomics[J]. Trends Biochem Sci, 2022, 47(3): 189-193.
doi: 10.1016/j.tibs.2021.11.002 URL |
| [7] |
Zhang Y, Zeng LR. Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity[J]. Plant Commun, 2020, 1(4): 100041.
doi: 10.1016/j.xplc.2020.100041 URL |
| [8] |
Chen YM, Hoehenwarter W. Changes in the phosphoproteome and metabolome link early signaling events to rearrangement of photosynthesis and central metabolism in salinity and oxidative stress response in Arabidopsis[J]. Plant Physiol, 2015, 169(4): 3021-3033.
doi: 10.1104/pp.15.01486 URL |
| [9] |
Barbour H, Nkwe NS, Estavoyer B, et al. An inventory of crosstalk between ubiquitination and other post-translational modifications in orchestrating cellular processes[J]. iScience, 2023, 26(5): 106276.
doi: 10.1016/j.isci.2023.106276 URL |
| [10] |
Wang X, Ding YL, Li ZY, et al. PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15[J]. Dev Cell, 2019, 51(2): 222-235.e5.
doi: S1534-5807(19)30691-4 pmid: 31543444 |
| [11] |
Liu JH, Wu XY, Fang Y, et al. A plant RNA virus inhibits NPR1 sumoylation and subverts NPR1-mediated plant immunity[J]. Nat Commun, 2023, 14(1): 3580.
doi: 10.1038/s41467-023-39254-2 pmid: 37328517 |
| [12] | 高丰衣, 陈艳梅. 质谱技术探索蛋白磷酸化信号机制的研究进展[J]. 科学通报, 2021, 66(20): 2529-2541. |
|
Gao FY, Chen YM. Decoding protein phosphorylation signaling networks by mass spectrometry[J]. Chin Sci Bull, 2021, 66(20): 2529-2541.
doi: 10.1360/TB-2020-1088 URL |
|
| [13] |
Chen XX, Ding YL, Yang YQ, et al. Protein kinases in plant responses to drought, salt, and cold stress[J]. J Integr Plant Biol, 2021, 63(1): 53-78.
doi: 10.1111/jipb.13061 |
| [14] |
Bhaskara GB, Wong MM, Verslues PE. The flip side of phospho-signalling: regulation of protein dephosphorylation and the protein phosphatase 2Cs[J]. Plant Cell Environ, 2019, 42(10): 2913-2930.
doi: 10.1111/pce.13616 |
| [15] |
Linden KJ, Callis J. The ubiquitin system affects agronomic plant traits[J]. J Biol Chem, 2020, 295(40): 13940-13955.
doi: 10.1074/jbc.REV120.011303 URL |
| [16] |
Filipčík P, Curry JR, Mace PD. When worlds collide-mechanisms at the interface between phosphorylation and ubiquitination[J]. J Mol Biol, 2017, 429(8): 1097-1113.
doi: S0022-2836(17)30086-4 pmid: 28235544 |
| [17] |
Dubeaux G, Neveu J, Zelazny E, et al. Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition[J]. Mol Cell, 2018, 69(6): 953-964.e5.
doi: S1097-2765(18)30107-2 pmid: 29547723 |
| [18] |
Dong J, Ni WM, Yu RB, et al. Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis[J]. Curr Biol, 2017, 27(16): 2420-2430.e6.
doi: S0960-9822(17)30799-6 pmid: 28736168 |
| [19] |
Yasuda S, Sato T, Maekawa S, et al. Phosphorylation of Arabidopsis ubiquitin ligase ATL31 is critical for plant carbon/nitrogen nutrient balance response and controls the stability of 14-3-3 proteins[J]. J Biol Chem, 2014, 289(22): 15179-15193.
doi: 10.1074/jbc.M113.533133 URL |
| [20] |
Sato T, Maekawa S, Yasuda S, et al. Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis[J]. Plant J, 2011, 68(1): 137-146.
doi: 10.1111/tpj.2011.68.issue-1 URL |
| [21] |
Yasuda S, Aoyama S, Hasegawa Y, et al. Arabidopsis CBL-interacting protein kinases regulate carbon/nitrogen-nutrient response by phosphorylating ubiquitin ligase ATL31[J]. Mol Plant, 2017, 10(4): 605-618.
doi: 10.1016/j.molp.2017.01.005 URL |
| [22] |
Nemoto K, Ramadan A, Arimura GI, et al. Tyrosine phosphorylation of the GARU E3 ubiquitin ligase promotes gibberellin signalling by preventing GID1 degradation[J]. Nat Commun, 2017, 8(1): 1004.
doi: 10.1038/s41467-017-01005-5 pmid: 29042542 |
| [23] |
Furlan G, Nakagami H, Eschen-Lippold L, et al. Changes in PUB22 ubiquitination modes triggered by MITOGEN-ACTIVATED PROTEIN KINASE3 dampen the immune response[J]. Plant Cell, 2017, 29(4): 726-745.
doi: 10.1105/tpc.16.00654 URL |
| [24] |
Wang J, Qu BY, Dou SJ, et al. The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity[J]. BMC Plant Biol, 2015, 15: 49.
doi: 10.1186/s12870-015-0442-4 pmid: 25849162 |
| [25] |
Metzger MB, Pruneda JN, Klevit RE, et al. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination[J]. Biochim Biophys Acta, 2014, 1843(1): 47-60.
doi: 10.1016/j.bbamcr.2013.05.026 pmid: 23747565 |
| [26] | Wang ZB, Stockinger E. Arabidopsis homologs of transcription-domain-associated protein 1(Tra1)are essential for embryogenesis, growth and development[J]. Dev Biol, 2006, 295(1): 384. |
| [27] |
Liu HX, Stone SL. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation[J]. Plant Cell, 2010, 22(8): 2630-2641.
doi: 10.1105/tpc.110.076075 URL |
| [28] |
Lyzenga WJ, Sullivan V, Liu HX, et al. The kinase activity of calcineurin B-like interacting protein kinase 26(CIPK26)influences its own stability and that of the ABA-regulated ubiquitin ligase, keep on going(KEG)[J]. Front Plant Sci, 2017, 8: 502.
doi: 10.3389/fpls.2017.00502 pmid: 28443108 |
| [29] |
Lu DP, Lin WW, Gao XQ, et al. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity[J]. Science, 2011, 332(6036): 1439-1442.
doi: 10.1126/science.1204903 pmid: 21680842 |
| [30] | Zhou JG, Liu DR, Wang P, et al. Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination[J]. Proc Natl Acad Sci USA, 2018, 115(8): E1906-E1915. |
| [31] |
Cheng CH, Wang ZJ, Ren ZY, et al. SCFAtPP2-B11 modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana[J]. PLoS Genet, 2017, 13(8): e1006947.
doi: 10.1371/journal.pgen.1006947 URL |
| [32] |
Niu D, Lin XL, Kong XX, et al. SIZ1-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis[J]. Mol Plant, 2019, 12(2): 215-228.
doi: S1674-2052(18)30368-X pmid: 30543996 |
| [33] |
Shahpasandzadeh H, Popova B, Kleinknecht A, et al. Interplay between sumoylation and phosphorylation for protection against α-synuclein inclusions[J]. J Biol Chem, 2014, 289(45): 31224-31240.
doi: 10.1074/jbc.M114.559237 pmid: 25231978 |
| [34] |
Saleh A, Withers J, Mohan R, et al. Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses[J]. Cell Host Microbe, 2015, 18(2): 169-182.
doi: 10.1016/j.chom.2015.07.005 pmid: 26269953 |
| [35] |
Lee HJ, Park YJ, Seo PJ, et al. Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in Arabidopsis[J]. Plant Cell, 2015, 27(12): 3425-3438.
doi: 10.1105/tpc.15.00371 URL |
| [36] |
Roy D, Sadanandom A. SUMO mediated regulation of transcription factors as a mechanism for transducing environmental cues into cellular signaling in plants[J]. Cell Mol Life Sci, 2021, 78(6): 2641-2664.
doi: 10.1007/s00018-020-03723-4 pmid: 33452901 |
| [37] |
Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis[J]. Nat Rev Mol Cell Biol, 2009, 10(8): 564-568.
doi: 10.1038/nrm2707 |
| [38] |
Conti L, Nelis S, Zhang CJ, et al. Small Ubiquitin-like Modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin[J]. Dev Cell, 2014, 28(1): 102-110.
doi: 10.1016/j.devcel.2013.12.004 pmid: 24434138 |
| [39] |
Guo RK, Sun WN. Sumoylation stabilizes RACK1B and enhance its interaction with RAP2.6 in the abscisic acid response[J]. Sci Rep, 2017, 7: 44090.
doi: 10.1038/srep44090 pmid: 28272518 |
| [40] |
Praefcke GJK, Hofmann K, Dohmen RJ. SUMO playing tag with ubiquitin[J]. Trends Biochem Sci, 2012, 37(1): 23-31.
doi: 10.1016/j.tibs.2011.09.002 pmid: 22018829 |
| [41] |
McManus FP, Lamoliatte F, Thibault P. Identification of cross talk between SUMOylation and ubiquitylation using a sequential peptide immunopurification approach[J]. Nat Protoc, 2017, 12(11): 2342-2358.
doi: 10.1038/nprot.2017.105 pmid: 29048423 |
| [42] |
Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin[J]. Trends Biochem Sci, 2008, 33(5): 201-208.
doi: 10.1016/j.tibs.2008.02.001 pmid: 18403209 |
| [43] |
Elrouby N, Bonequi MV, Porri A, et al. Identification of Arabidopsis SUMO-interacting proteins that regulate chromatin activity and developmental transitions[J]. Proc Natl Acad Sci USA, 2013, 110(49): 19956-19961.
doi: 10.1073/pnas.1319985110 pmid: 24255109 |
| [44] |
Budhiraja R, Hermkes R, Müller S, et al. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation[J]. Plant Physiol, 2009, 149(3): 1529-1540.
doi: 10.1104/pp.108.135053 pmid: 19151129 |
| [45] |
Lin XL, Niu D, Hu ZL, et al. An Arabidopsis SUMO E3 ligase, SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity[J]. PLoS Genet, 2016, 12(4): e1006016.
doi: 10.1371/journal.pgen.1006016 URL |
| [46] |
Huang TT, Wuerzberger-Davis SM, Wu ZH, et al. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress[J]. Cell, 2003, 115(5): 565-576.
doi: 10.1016/s0092-8674(03)00895-x pmid: 14651848 |
| [47] |
Sadanandom A, Ádám É, Orosa B, et al. SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana[J]. Proc Natl Acad Sci USA, 2015, 112(35): 11108-11113.
doi: 10.1073/pnas.1415260112 pmid: 26283376 |
| [48] |
Crozet P, Margalha L, Butowt R, et al. SUMOylation represses SnRK1 signaling in Arabidopsis[J]. Plant J, 2016, 85(1): 120-133.
doi: 10.1111/tpj.2016.85.issue-1 URL |
| [49] |
Kim JY, Park BS, Park SW, et al. Nitrate reductases are relocalized to the nucleus by AtSIZ1 and their levels are negatively regulated by COP1 and ammonium[J]. Int J Mol Sci, 2018, 19(4): 1202.
doi: 10.3390/ijms19041202 URL |
| [50] |
Lambeck IC, Fischer-Schrader K, Niks D, et al. Molecular mechanism of 14-3-3 protein-mediated inhibition of plant nitrate reductase[J]. J Biol Chem, 2012, 287(7): 4562-4571.
doi: 10.1074/jbc.M111.323113 pmid: 22170050 |
| [51] |
Heazlewood JL, Durek P, Hummel J, et al. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor[J]. Nucleic Acids Res, 2008, 36(Database issue): D1015-D1021.
doi: 10.1093/nar/gkm812 pmid: 17984086 |
| [52] |
Durek P, Schmidt R, Heazlewood JL, et al. PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update[J]. Nucleic Acids Res, 2010, 38(Database issue): D828-D834.
doi: 10.1093/nar/gkp810 URL |
| [53] |
Chen YM, Wang Y, Liang XL, et al. Mass spectrometric exploration of phytohormone profiles and signaling networks[J]. Trends Plant Sci, 2023, 28(4): 399-414.
doi: 10.1016/j.tplants.2022.12.006 URL |
| [54] |
Chen YM, Liu P. Proteome-wide chromatin interactomics to study plant epigenetics[J]. Trends Plant Sci, 2021, 26(7): 758-759.
doi: 10.1016/j.tplants.2021.04.002 URL |
| [1] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [2] | LU Yu-dan, LIU Xiao-chi, FENG Xin, CHEN Gui-xin, CHEN Yi-ting. Identification of the Kiwifruit BBX Gene Family and Analysis of Their Transcriptional Characteristics [J]. Biotechnology Bulletin, 2024, 40(2): 172-182. |
| [3] | LI Ya-nan, ZHANG Hao-jie, LIANG Meng-jing, LUO Tao, LI Wang-ning, ZHANG Chun-hui, JI Chun-li, LI Run-zhi, XUE Jin-ai, CUI Hong-li. Identification and Expression Analysis of Calcium-dependent Protein Kinase(CDPK)Family in Haematococcus pluvialis [J]. Biotechnology Bulletin, 2024, 40(2): 300-312. |
| [4] | ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses [J]. Biotechnology Bulletin, 2024, 40(2): 90-98. |
| [5] | ZHOU Hui-wen, WU Lan-hua, HAN De-peng, ZHENG Wei, YU Pao-lan, WU Yang, XIAO Xiao-jun. Genome-wide Association Study of Seed Glucosinolate Content in Brassica napus [J]. Biotechnology Bulletin, 2024, 40(1): 222-230. |
| [6] | WU Zhen, ZHANG Ming-Ying, YAN Feng, LI Yi-min, GAO Jing, YAN Yong-Gang, ZHANG Gang. Identification and Analysis of WRKY Gene Family in Rheum palmatum L. [J]. Biotechnology Bulletin, 2024, 40(1): 250-261. |
| [7] | WANG Bin, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui, HE Jin-ming. Cloning of bHLH96 Gene and Its Roles in Regulating the Biosynthesis of Peppermint Terpenes [J]. Biotechnology Bulletin, 2024, 40(1): 281-293. |
| [8] | CHEN Zhi-min, LI Cui, WEI Ji-tian, LI Xin-ran, LIU Yi, GUO Qiang. Research Progress in the Regulation of Chlorogenic Acid Biosynthesis and Its Application [J]. Biotechnology Bulletin, 2024, 40(1): 57-71. |
| [9] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [10] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [11] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [12] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [13] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [14] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [15] | HU Hai-lin, XU Li, LI Xiao-xu, WANG Chen-can, MEI Man, DING Wen-jing, ZHAO Yuan-yuan. Advances in the Regulation of Plant Growth, Development and Stress Physiology by Small Peptide Hormones [J]. Biotechnology Bulletin, 2023, 39(7): 13-25. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
