








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (2): 289-299.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0877
Previous Articles Next Articles
ZHENG Fei( ), YANG Jun-zhao, NIU Yu-feng, LI Rui-lin, ZHAO Guo-zhu(
), YANG Jun-zhao, NIU Yu-feng, LI Rui-lin, ZHAO Guo-zhu( )
)
Received:2023-09-12
Online:2024-02-26
Published:2024-03-13
Contact:
ZHAO Guo-zhu
E-mail:zhengfei0718@bjfu.edu.cn;zhaogz@bjfu.edu.cn
ZHENG Fei, YANG Jun-zhao, NIU Yu-feng, LI Rui-lin, ZHAO Guo-zhu. Characterization and Functional Analysis of Lytic Polysaccharide Monooxygenase TtLPMO9I from Thermothelomyces thermophilus[J]. Biotechnology Bulletin, 2024, 40(2): 289-299.
| 序号No. | 蛋白 Protein | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 蛋白结构域 Protein domain | 参考文献 Reference |
|---|---|---|---|---|---|
| 1 | MYCTH_112089 | 232 | 26.1 |  | [ |
| 2 | MtLPMO9B | 323 | 32.4 |  | [ |
| 3 | MtLPMO9D | 255 | 27.0 |  | [ |
| 4 | MtLPMO9A | 225 | 24.4 |  | [ |
| 5 | AEO59955.1 | 235 | 24.4 |  | 未报道 |
| 6 | AEO54509.1 | 303 | 31.5 |  | 未报道 |
| 7 | MtLPMO9 | 342 | 34.9 |  | [ |
| 8 | TtLPMO9I | 306 | 31.4 |  | 本文研究材料 |
| 9 | MtLPMO9J | 246 | 26.0 |  | [ |
| 10 | AEO59836.1 | 227 | 24.1 |  | 未报道 |
| 11 | AEO59482.1 | 198 | 21.5 |  | 未报道 |
| 12 | AEO58921.1 | 340 | 35.8 |  | 未报道 |
| 13 | AEO58412.1 | 338 | 35.2 |  | 未报道 |
| 14 | AEO61257.1 | 241 | 25.9 |  | 未报道 |
| 15 | AEO56498.1a | 151 | 16.2 |  | 未报道 |
| 16 | AEO59823.1 | 254 | 28.4 |  | 未报道 |
| 17 | AEO55776.1 | 444 | 46.6 |  | 未报道 |
| 18 | AEO61305.1 | 230 | 25.0 |  | 未报道 |
| 19 | AEO55082.1 | 226 | 24.5 |  | 未报道 |
| 20 | MtLPMO9C | 237 | 24.9 |  | [ |
| 21 | AEO56547.1 | 245 | 26.5 |  | 未报道 |
| 22 | AEO55652.1 | 242 | 25.5 |  | 未报道 |
Table 1 Protein property prediction of the LPMO AA9 family from T. thermophilus
| 序号No. | 蛋白 Protein | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 蛋白结构域 Protein domain | 参考文献 Reference |
|---|---|---|---|---|---|
| 1 | MYCTH_112089 | 232 | 26.1 |  | [ |
| 2 | MtLPMO9B | 323 | 32.4 |  | [ |
| 3 | MtLPMO9D | 255 | 27.0 |  | [ |
| 4 | MtLPMO9A | 225 | 24.4 |  | [ |
| 5 | AEO59955.1 | 235 | 24.4 |  | 未报道 |
| 6 | AEO54509.1 | 303 | 31.5 |  | 未报道 |
| 7 | MtLPMO9 | 342 | 34.9 |  | [ |
| 8 | TtLPMO9I | 306 | 31.4 |  | 本文研究材料 |
| 9 | MtLPMO9J | 246 | 26.0 |  | [ |
| 10 | AEO59836.1 | 227 | 24.1 |  | 未报道 |
| 11 | AEO59482.1 | 198 | 21.5 |  | 未报道 |
| 12 | AEO58921.1 | 340 | 35.8 |  | 未报道 |
| 13 | AEO58412.1 | 338 | 35.2 |  | 未报道 |
| 14 | AEO61257.1 | 241 | 25.9 |  | 未报道 |
| 15 | AEO56498.1a | 151 | 16.2 |  | 未报道 |
| 16 | AEO59823.1 | 254 | 28.4 |  | 未报道 |
| 17 | AEO55776.1 | 444 | 46.6 |  | 未报道 |
| 18 | AEO61305.1 | 230 | 25.0 |  | 未报道 |
| 19 | AEO55082.1 | 226 | 24.5 |  | 未报道 |
| 20 | MtLPMO9C | 237 | 24.9 |  | [ |
| 21 | AEO56547.1 | 245 | 26.5 |  | 未报道 |
| 22 | AEO55652.1 | 242 | 25.5 |  | 未报道 |
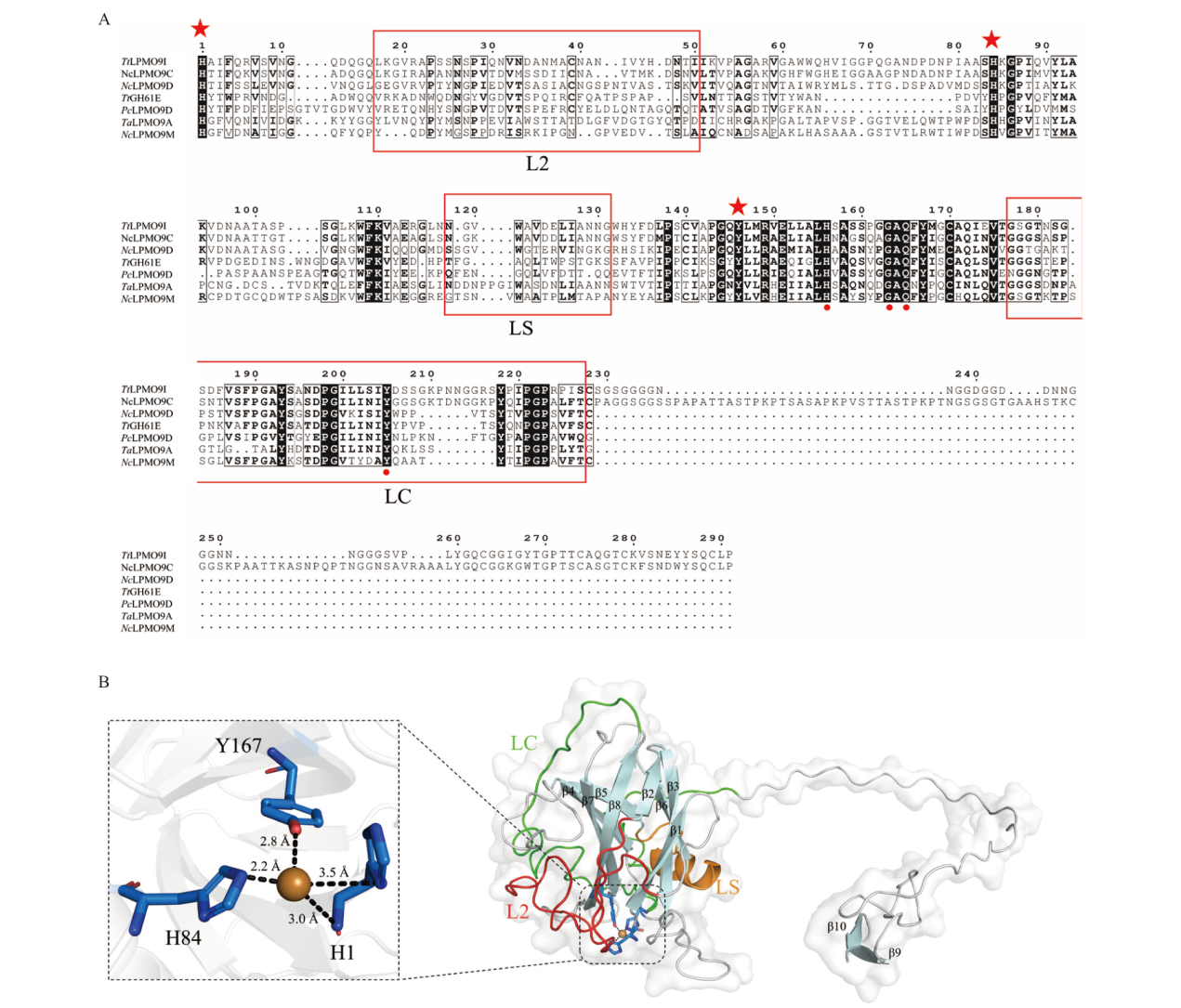
Fig. 2 Bioinformatics analysis of TtLPMO9I A: Multiple sequence alignment of TtLPMO9I with LPMO in crystal structure. B: 3D structural simulation diagram of TtLPMO9I
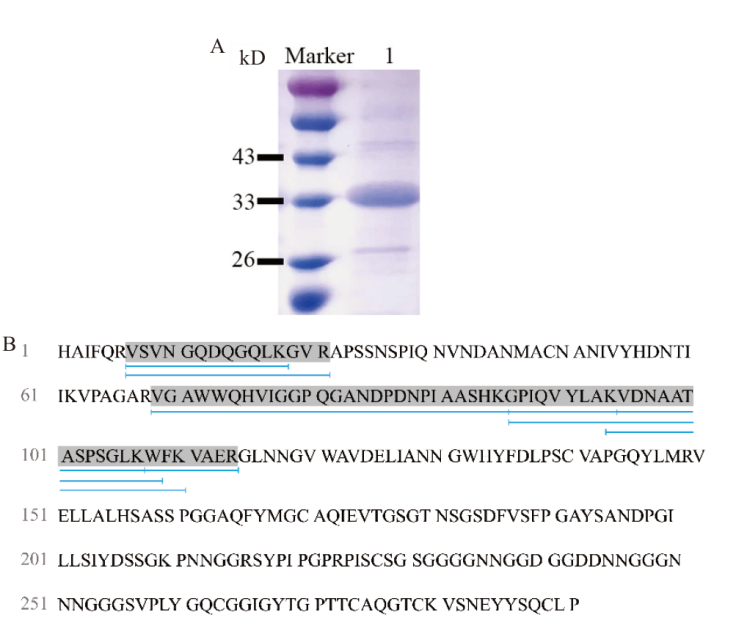
Fig. 3 Validation of the recombinant protein TtLPMO9I A: SDS-PAGE analysis of TtLPMO9I. B: MALDI-TOF mass spectrometry analysis results diagram for TtLPMO9I. Marker: Protein molecular weight standard. 1: Purified protein of TtLPMO9I

Fig. 5 Enzymatic properties of TtLPMO9I A: Effect of temperature on the cellulase activity of TtLPMO9I. B: Effect of temperature on the stability of TtLPMO9I. C: Effect of pH on the cellulase activity of TtLPMO9I. D: Effect of pH on the viability stability of TtLPMO9I

Fig. 6 Synergistic reaction between TtLPMO9I and cellulase(Novozymes 188)for the degradation of pretreated corn straw and Avicel A: Pretreated corn straw was used as substrate for 12 h. B: Pretreated corn straw was used as substrate for 24 h. C: Reaction with Avicel as substrate for 12 h. D: Reaction with Avicel as substrate for 24 h. The blue block in the figure above indicates the amount of reducing sugar produced by adding 50, 100, and 200 μg TtLPMO9I, respectively. The green block indicates the amount of reducing sugar produced by adding 100 μL cellulase. The orange block indicates the amount of reducing sugar produced by 50 μg TtLPMO9I+100 μL cellulase, 100 μg TtLPMO9I+100 μL cellulase, and 200 μg TtLPMO9I+100 μL cellulase
| [1] | 宫秀杰, 钱春荣, 于洋, 等. 近年纤维素降解菌株筛选研究进展[J]. 纤维素科学与技术, 2021, 29(2): 68-77. |
| Gong XJ, Qian CR, Yu Y, et al. Progress on screening of cellulose degrading strains in recent years[J]. J Cellul Sci Technol, 2021, 29(2): 68-77. | |
| [2] |
Levasseur A, Drula E, Lombard V, et al. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes[J]. Biotechnol Biofuels, 2013, 6(1): 41.
doi: 10.1186/1754-6834-6-41 pmid: 23514094 |
| [3] |
Karkehabadi S, Hansson H, et al. The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 A resolution[J]. J Mol Biol, 2008, 383(1): 144-154.
doi: 10.1016/j.jmb.2008.08.016 pmid: 18723026 |
| [4] | 李欣, 张丽丽, 田莉, 等. 裂解多糖单加氧酶高效催化的研究进展[J]. 生物化学与生物物理进展, 2016, 43(10): 970-979. |
| Li X, Zhang LL, Tian L, et al. The advance of efficient catalysis of lytic polysaccharide monooxygenases[J]. Prog Biochem Biophys, 2016, 43(10): 970-979. | |
| [5] |
Sun PC, Valenzuela SV, et al. Oxidized product profiles of AA9 lytic polysaccharide monooxygenases depend on the type of cellulose[J]. ACS Sustain Chem Eng, 2021, 9(42): 14124-14133.
doi: 10.1021/acssuschemeng.1c04100 pmid: 34722005 |
| [6] |
Couturier M, Ladevèze S, Sulzenbacher G, et al. Lytic xylan oxidases from wood-decay fungi unlock biomass degradation[J]. Nat Chem Biol, 2018, 14(3): 306-310.
doi: 10.1038/nchembio.2558 pmid: 29377002 |
| [7] |
Sabbadin F, Hemsworth GR, Ciano L, et al. An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion[J]. Nat Commun, 2018, 9(1): 756.
doi: 10.1038/s41467-018-03142-x pmid: 29472725 |
| [8] |
Filiatrault-Chastel C, Navarro D, Haon M, et al. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes[J]. Biotechnol Biofuels, 2019, 12: 55.
doi: 10.1186/s13068-019-1394-y pmid: 30923563 |
| [9] |
Sabbadin F, Urresti S, Henrissat B, et al. Secreted pectin monooxygenases drive plant infection by pathogenic oomycetes[J]. Science, 2021, 373(6556): 774-779.
doi: 10.1126/science.abj1342 pmid: 34385392 |
| [10] |
Calderaro F, Keser M, Akeroyd M, et al. Characterization of an AA9 LPMO from Thielavia australiensis, TausLPMO9B, under industrially relevant lignocellulose saccharification conditions[J]. Biotechnol Biofuels, 2020, 13(1): 195.
doi: 10.1186/s13068-020-01836-3 pmid: 33292403 |
| [11] | Hegnar OA, Petrovic DM, Bissaro B, et al. pH-dependent relationship between catalytic activity and hydrogen peroxide production shown via characterization of a lytic polysaccharide monooxygenase from Gloeophyllum trabeum[J]. Appl Environ Microbiol, 2019, 85(5): e02612-e02618. |
| [12] |
Agrawal D, Kaur B, Kaur Brar K, et al. An innovative approach of priming lignocellulosics with lytic polysaccharide mono-oxygenases prior to saccharification with glycosyl hydrolases can economize second generation ethanol process[J]. Bioresour Technol, 2020, 308: 123257.
doi: 10.1016/j.biortech.2020.123257 URL |
| [13] |
Müller G, Várnai A, Johansen KS, et al. Harnessing the potential of LPMO-containing cellulase cocktails poses new demands on processing conditions[J]. Biotechnol Biofuels, 2015, 8: 187.
doi: 10.1186/s13068-015-0376-y pmid: 26609322 |
| [14] | 吴帅帅, 田娟, 谢宁, 等. AA9家族裂解多糖单加氧酶研究进展[J]. 纤维素科学与技术, 2020, 28(3): 70-79. |
| Wu SS, Tian J, Xie N, et al. Progress of AA9 family lytic polysaccharide monooxygenase[J]. J Cellul Sci Technol, 2020, 28(3): 70-79. | |
| [15] |
Frommhagen M, Westphal AH, et al. Distinct substrate specificities and electron-donating systems of fungal lytic polysaccharide monooxygenases[J]. Front Microbiol, 2018, 9: 1080.
doi: 10.3389/fmicb.2018.01080 pmid: 29896168 |
| [16] |
Chen C, Chen JY, Geng ZG, et al. Regioselectivity of oxidation by a polysaccharide monooxygenase from Chaetomium thermophi-lum[J]. Biotechnol Biofuels, 2018, 11: 155.
doi: 10.1186/s13068-018-1156-2 pmid: 29991963 |
| [17] |
Beeson WT, Vu VV, Span EA, et al. Cellulose degradation by polysaccharide monooxygenases[J]. Annu Rev Biochem, 2015, 84: 923-946.
doi: 10.1146/annurev-biochem-060614-034439 pmid: 25784051 |
| [18] |
Danneels B, Tanghe M, Joosten HJ, et al. A quantitative indicator diagram for lytic polysaccharide monooxygenases reveals the role of aromatic surface residues in HjLPMO9A regioselectivity[J]. PLoS One, 2017, 12(5): e0178446.
doi: 10.1371/journal.pone.0178446 URL |
| [19] |
Vu VV, Beeson WT, et al. Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases[J]. J Am Chem Soc, 2014, 136(2): 562-565.
doi: 10.1021/ja409384b pmid: 24350607 |
| [20] |
Frommhagen M, Koetsier MJ, Westphal AH, et al. Lytic polysaccharide monooxygenases from Myceliophthora thermophila C1 differ in substrate preference and reducing agent specificity[J]. Biotechnol Biofuels, 2016, 9(1): 186.
doi: 10.1186/s13068-016-0594-y pmid: 27588039 |
| [21] |
Frommhagen M, Westphal AH, Hilgers R, et al. Quantification of the catalytic performance of C1-cellulose-specific lytic polysaccharide monooxygenases[J]. Appl Microbiol Biotechnol, 2018, 102(3): 1281-1295.
doi: 10.1007/s00253-017-8541-9 pmid: 29196788 |
| [22] |
Frommhagen M, Sforza S, Westphal AH, et al. Discovery of the combined oxidative cleavage of plant xylan and cellulose by a new fungal polysaccharide monooxygenase[J]. Biotechnol Biofuels, 2015, 8: 101.
doi: 10.1186/s13068-015-0284-1 pmid: 26185526 |
| [23] |
Karnaouri A, Muraleedharan MN, Dimarogona M, et al. Recombinant expression of thermostable processive Mt EG5 endoglucanase and its synergism with Mt LPMO from Myceliophthora thermophila during the hydrolysis of lignocellulosic substrates[J]. Biotechnol Biofuels, 2017, 10: 126.
doi: 10.1186/s13068-017-0813-1 pmid: 28515785 |
| [24] |
Kadowaki MAS, Várnai A, Jameson JK, et al. Functional characterization of a lytic polysaccharide monooxygenase from the thermophilic fungus Myceliophthora thermophila[J]. PLoS One, 2018, 13(8): e0202148.
doi: 10.1371/journal.pone.0202148 URL |
| [25] |
Hangasky JA, Marletta MA. A random-sequential kinetic mechanism for polysaccharide monooxygenases[J]. Biochemistry, 2018, 57(22): 3191-3199.
doi: 10.1021/acs.biochem.8b00129 pmid: 29683313 |
| [26] |
Berka RM, Grigoriev IV, Otillar R, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Mycelioph-thora thermophila and Thielavia terrestris[J]. Nat Biotechnol, 2011, 29(10): 922-927.
doi: 10.1038/nbt.1976 |
| [27] |
Galagan JE, Calvo SE, Cuomo C, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. ory-zae[J]. Nature, 2005, 438(7071): 1105-1115.
doi: 10.1038/nature04341 |
| [28] |
Espagne E, Lespinet O, Malagnac F, et al. The genome sequence of the model ascomycete fungus Podospora anserina[J]. Genome Biol, 2008, 9(5): R77.
doi: 10.1186/gb-2008-9-5-r77 URL |
| [29] |
Galagan JE, Calvo SE, Borkovich KA, et al. The genome sequence of the filamentous fungus Neurospora crassa[J]. Nature, 2003, 422(6934): 859-868.
doi: 10.1038/nature01554 URL |
| [30] |
Book AJ, Yennamalli RM, Takasuka TE, et al. Evolution of substrate specificity in bacterial AA10 lytic polysaccharide monooxygenases[J]. Biotechnol Biofuels, 2014, 7: 109.
doi: 10.1186/1754-6834-7-109 pmid: 25161697 |
| [31] |
Hervé C, Rogowski A, Blake AW, et al. Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects[J]. Proc Natl Acad Sci USA, 2010, 107(34): 15293-15298.
doi: 10.1073/pnas.1005732107 pmid: 20696902 |
| [32] |
Forsberg Z, Courtade G. On the impact of carbohydrate-binding modules(CBMs)in lytic polysaccharide monooxygenases(LPMOs)[J]. Essays Biochem, 2023, 67(3): 561-574.
doi: 10.1042/EBC20220162 URL |
| [33] |
马玉倩, 孙东辉, 岳浩峰, 等. 具有辅助降解纤维素功能的大斑刚毛座腔菌糖苷水解酶GH61的鉴定、异源表达及功能分析[J]. 生物技术通报, 2023, 39(4): 124-135.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0996 |
| Ma YQ, Sun DH, et al. Identification, heterologous expression and functional analysis of a GH61 family glycoside hydrolase from Setosphaeria turcica with the assisting function in degrading cellulose[J]. Biotechnol Bull, 2023, 39(4): 124-135. | |
| [34] |
Li YL, Li TB, Guo JT, et al. Expression and characterization of a novel lytic polysaccharide monooxygenase, PdLPMO9A, from the edible fungus Pleurotus djamor and its synergistic interactions with cellulase in corn straw biomass saccharification[J]. Bioresour Technol, 2022, 348: 126792.
doi: 10.1016/j.biortech.2022.126792 URL |
| [35] |
Srivastava A, Nagar P, Rathore S, et al. The linker region promotes activity and binding efficiency of modular LPMO towards polymeric substrate[J]. Microbiol Spectr, 2022, 10(1): e0269721.
doi: 10.1128/spectrum.02697-21 URL |
| [36] |
Dimarogona M, Topakas E, Olsson L, et al. Lignin boosts the cellulase performance of a GH-61 enzyme from Sporotrichum thermo-phile[J]. Bioresour Technol, 2012, 110: 480-487.
doi: 10.1016/j.biortech.2012.01.116 URL |
| [37] | Bulakhov AG, Gusakov AV, Chekushina AV, et al. Isolation of homogeneous polysaccharide monooxygenases from fungal sources and investigation of their synergism with cellulases when acting on cellulose[J]. Biochemistry, 2016, 81(5): 530-537. |
| [38] |
Harris PV, Welner D, McFarland KC, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family[J]. Biochemistry, 2010, 49(15): 3305-3316.
doi: 10.1021/bi100009p pmid: 20230050 |
| [39] |
Rodríguez-Zúñiga UF, Cannella D, de Campos Giordano R, et al. Lignocellulose pretreatment technologies affect the level of enzymatic cellulose oxidation by LPMO[J]. Green Chem, 2015, 17(5): 2896-2903.
doi: 10.1039/C4GC02179G URL |
| [1] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [2] | ZHAO Sai-sai, ZHANG Xiao-dan, JIA Xiao-yan, TAO Da-wei, LIU Ke-yu, NING Xi-bin. Investigation on the Complex Mutagenesis Selection of High-yield Nitrate Reductase Strain Staphylococcus simulans ZSJ6 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2023, 39(4): 103-113. |
| [3] | MA Yu-qian, SUN Dong-hui, YUE Hao-feng, XIN Jia-yu, LIU Ning, CAO Zhi-yan. Identification, Heterologous Expression and Functional Analysis of a GH61 Family Glycoside Hydrolase from Setosphaeria turcica with the Assisting Function in Degrading Cellulose [J]. Biotechnology Bulletin, 2023, 39(4): 124-135. |
| [4] | CHEN Nan-nan, WANG Chun-lai, JIANG Zhen-zhong, JIAO Peng, GUAN Shu-yan, MA Yi-yong. Genetic Transformation and Chilling Resistance Analysis of Maize ZmDHN15 Gene in Tobacco [J]. Biotechnology Bulletin, 2023, 39(4): 259-267. |
| [5] | ZHANG Kai-ping, LIU Yan-li, TU Mian-liang, LI Ji-wei, WU Wen-biao. Optimization of Producing Cellulase by Aspergillus fumigatus A-16 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2022, 38(9): 215-225. |
| [6] | NIU Xin, ZHANG Ying, WANG Mao-jun, LIU Wen-long, LU Fu-ping, LI Yu. Effects of Different Integration Sites on the Expression of Exogenous Alkaline Protease in Bacillus amyloliquefaciens [J]. Biotechnology Bulletin, 2022, 38(4): 253-260. |
| [7] | CHANG Qing, SHU Yue-rong, WANG Wen-tao, JIANG Hao, YAN Quan-de, QIAN Zheng, GAO Xue-chun, WU Jin-hong, ZHANG Yong. Heterologous Expression and Characterization of Endo-type Alginate Lyase from Yeosuana marina sp. JLT21 [J]. Biotechnology Bulletin, 2022, 38(2): 123-131. |
| [8] | WANG Bo-ya, JIANG Yong, HUANG Yan, CAO Ying, HU Shang-lian. Cloning and Functional Analysis of BeCesA4 in Bambusa emeiensis [J]. Biotechnology Bulletin, 2022, 38(11): 185-193. |
| [9] | WANG Xiao-tao, ZOU Hang, WU Yi, XIANG Shen-wei, LV Hua, LIU Chao-lan, LIN Jia-fu, WANG Xin-rong, CHU Yi-wen, SONG Tao. Heterologous Expression and Enzymatic Properties Analysis of Novel β-agarase Aga2 from Paraglaciecola hydrolytica [J]. Biotechnology Bulletin, 2022, 38(11): 258-268. |
| [10] | ZHANG Tong-tong, ZHENG Deng-yu, WU Zhong-yi, ZHANG Zhong-bao, YU Rong. Functional Analysis of ZmNF-YB13 Responding to Drought and Salt Stress [J]. Biotechnology Bulletin, 2022, 38(10): 115-123. |
| [11] | TIAN Jia-hui, FENG Jia-li, LU Jun-hua, MAO Lin-jing, HU Zhu-ran, WANG Ying, CHU Jie. Isolation,Purification and Characterization of Laccase LacT-1 from Cerrena unicolor [J]. Biotechnology Bulletin, 2021, 37(8): 186-194. |
| [12] | LIAO Zhao-min, CAI Jun, LIN Jian-guo, DU Xin, WANG Chang-gao. Expression of Glucose Oxidase Gene from Aspergillus niger in Pichia pastoris and Optimization of Enzyme Production Conditions [J]. Biotechnology Bulletin, 2021, 37(6): 97-107. |
| [13] | LIU Shan, YE Wei, ZHU Mu-zi, LI Sai-ni, DENG Zhang-shuang, ZHANG Wei-min. Cloning,Expression and Characterization of a Novel Acyltransferase GPAT [J]. Biotechnology Bulletin, 2021, 37(11): 257-266. |
| [14] | ZHAO Hai-yan, SONG Chen-bin, LIU Zheng-ya, MA Xing-rong, SHANG Hui-hui, LI An-hua, GUAN Xian-jun, WANG Jian-she. Cloning,Recombinant Expression and Enzymatic Properties of α-Amylase Gene from Laceyella sp. [J]. Biotechnology Bulletin, 2020, 36(8): 23-33. |
| [15] | LI Wei-na, SHEN Dong-ling, ZHANG Yu-xing, LIU Xue-tong, IRBIS Chagan. Cloning and Enzymatic Identification of Thermo-tolerant and Endotype Alginate Lyase Gene from Mangrovibacterium sp. SH-52 [J]. Biotechnology Bulletin, 2020, 36(12): 82-90. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||