








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (2): 123-131.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0539
Previous Articles Next Articles
CHANG Qing1( ), SHU Yue-rong1, WANG Wen-tao1, JIANG Hao1, YAN Quan-de1, QIAN Zheng1, GAO Xue-chun1, WU Jin-hong2, ZHANG Yong1(
), SHU Yue-rong1, WANG Wen-tao1, JIANG Hao1, YAN Quan-de1, QIAN Zheng1, GAO Xue-chun1, WU Jin-hong2, ZHANG Yong1( )
)
Received:2021-04-22
Online:2022-02-26
Published:2022-03-09
Contact:
ZHANG Yong
E-mail:19921870965@163.com;yzhang2011@sjtu.edu.cn
CHANG Qing, SHU Yue-rong, WANG Wen-tao, JIANG Hao, YAN Quan-de, QIAN Zheng, GAO Xue-chun, WU Jin-hong, ZHANG Yong. Heterologous Expression and Characterization of Endo-type Alginate Lyase from Yeosuana marina sp. JLT21[J]. Biotechnology Bulletin, 2022, 38(2): 123-131.

Fig.1 Phylogenetic tree of alginate lyase YMA-1 and amino acid sequence alignment analysis of PL7 family enzyme A:Bootstraps are the confidence values obtained from 1,000 replications(%). Species of alginolytic bacteria and the accession number from GenBank are listed on the right. Scale bar indicates approximately 20% sequence difference. B:FlAlyA from Flavobacterium sp. UMI-01(BAP05660),A1-II’ from Sphingomonas sp. A1(BAD16656),AlyA from Klebsiella pneumoniae(AAA25049),AlyA5 from Zobellia galactanivorans DsijT(CAZ98266.1),AlyA1 from Z. galactanivorans DsiJT(CAZ95239),AlgAT5 from Defluviitalea phaphyphila(WP_058486006.1),PA1167 from Pseudomonas aeruginosa PAO1(AAG04556),and AlyC3 from Psychromonas sp.(QOP59290.1)

Fig.2 Recombinant expression,purification and analysis of alginate lyase YMA-1 A:Detecting the purity of recombinase YMA-1 via SDS-PAGE. M:Protein marker. 1:Crude enzyme. 2:Flow-through with 20 mmol/L imidazole buffer;3:Flow-through with 30 mM imidazole buffer. 4:Flow-through with 50 mmol/L imidazole buffer. 5:Flow-through with 80 mmol/L imidazole buffer. 6:Purified recombinant enzyme YMA-1. B:Gel filtration chromatography analysis of recombinant enzyme YMA-1
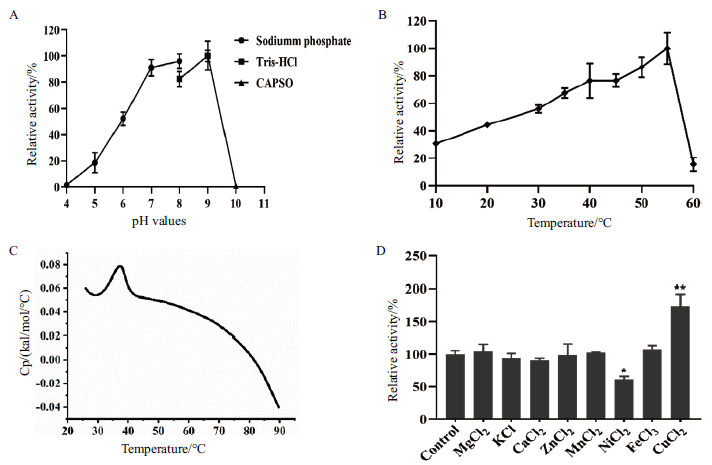
Fig.3 Activities of alginate lyase YMA-1 under different pH,temperature and metal ions A:Determination of YMA-1 activity at pH 5.0 to 10.0. YMA-1 catalysis was optimal at pH 9.0,and its specific activity of the enzyme was 4.7×103 U/mg. B:Examination of YMA-1 activity at 10℃ to 60℃. YMA-1 showed the optimal activity of 1.3×104 U/mg at 55℃. C:Measurement of YMA-1 thermostability Tm value was determined by differential scanning calorimetry(DSC). D:Effects of various metal ions on the activity of YMA-1

Fig.4 Substrate preference of alginate lyase YMA-1 A:Substrate specificity of alginate lyases. Alginate lyases can be classified into Poly M lyase and Poly G lyase and bi-functional lyase according to chemical structures of substrates acting with alginate lyase. B:Preference of YMA-1 on 3 substrates in different structures,it presents high activity on 3 alginic acid substrates in different structure,confirming it is a bi-functional alginate lyase

Fig.6 LC-MS analysis of the oligosaccharide products from degrading sodium alginates by YMA-1 catalysis A:HPLC analysis of products of YMA-1 catalysis. Four different end-products were detected,and the peak times were 3.92,4.04,4.13 and 4.23 min,respectively. B:ESI-MST analysis of products of YMA-1 catalysis. The end-products were unsaturated disaccharides(ΔDP2)([M-H]-=351.0),unsaturated trisaccharides(ΔDP3)([M-H]-=527.09),unsaturated tetrasaccharide(ΔDP4)([M-H]-=703.12)and unsaturated pentasaccharide(ΔDP5)([M-H]-=879.15),respectively
| [1] |
Kloareg B, Demarty M, Mabeau S. Ion-exchange properties of isolated cell walls of brown algae:the interstitial solution[J]. J Exp Bot, 1987, 38(10):1652-1662.
doi: 10.1093/jxb/38.10.1652 URL |
| [2] |
Sellimi S, Younes I, Ayed HB, et al. Structural, physicochemical and antioxidant properties of sodium alginate isolated from a Tunisian brown seaweed[J]. Int J Biol Macromol, 2015, 72:1358-1367.
doi: 10.1016/j.ijbiomac.2014.10.016 URL |
| [3] |
Tang JC, Taniguchi H, Chu H, et al. Isolation and characterization of alginate-degrading bacteria for disposal of seaweed wastes[J]. Lett Appl Microbiol, 2009, 48(1):38-43.
doi: 10.1111/j.1472-765X.2008.02481.x pmid: 19018967 |
| [4] |
Wong TY, Preston LA, Schiller NL. Alginate lyase:review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications[J]. Annu Rev Microbiol, 2000, 54(1):289-340.
doi: 10.1146/micro.2000.54.issue-1 URL |
| [5] |
Jain S, Ohman DE. Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa[J]. Infect Immun, 2005, 73(10):6429-6436.
doi: 10.1128/IAI.73.10.6429-6436.2005 URL |
| [6] |
Cheng D, Jiang C, Xu J, et al. Characteristics and applications of alginate lyases:a review[J]. Int J Biol Macromol, 2020, 164:1304-1320.
doi: 10.1016/j.ijbiomac.2020.07.199 URL |
| [7] |
Zhu B, Yin H. Alginate lyase:Review of major sources and classification, properties, structure-function analysis and applications[J]. Bioengineered, 2015, 6(3):125-131.
doi: 10.1080/21655979.2015.1030543 URL |
| [8] | Xu F, Wang P, Zhang YZ, et al. Diversity of three-dimensional structures and catalytic mechanisms of alginate lyases[J]. Appl Environ Microbiol, 2018, 84(3):e02040-17. |
| [9] |
Lombard V, Bernard T, Rancurel C, et al. A hierarchical classification of polysaccharide lyases for glycogenomics[J]. Biochem J, 2010, 432(3):437-444.
doi: 10.1042/BJ20101185 URL |
| [10] |
Wang B, Ji SQ, Lu M, et al. Biochemical and structural characterization of alginate lyases:an update[J]. Curr Biotechnol, 2015, 4(3):223-239.
doi: 10.2174/2211550104666150723231423 URL |
| [11] |
Nakata S, Murata K, Hashimoto W, et al. Uncovering the reactive nature of 4-deoxy- l - erythro -5-hexoseulose uronate for the utilization of alginate, a promising marine biopolymer[J]. Sci Rep, 2019, 9:17147.
doi: 10.1038/s41598-019-53597-1 pmid: 31748627 |
| [12] |
Kim HS, Lee CG, Lee EY. Alginate lyase:Structure, property, and application[J]. Biotechnol Bioprocess Eng, 2011, 16(5):843-851.
doi: 10.1007/s12257-011-0352-8 URL |
| [13] |
Wang Y, Han F, Hu B, et al. In vivo prebiotic properties of alginate oligosaccharides prepared through enzymatic hydrolysis of alginate[J]. Nutr Res, 2006, 26(11):597-603.
doi: 10.1016/j.nutres.2006.09.015 URL |
| [14] |
Islan GA, Bosio VE, Castro GR. Alginate lyase and ciprofloxacin co-immobilization on biopolymeric microspheres for cystic fibrosis treatment[J]. Macromol Biosci, 2013, 13(9):1238-1248.
doi: 10.1002/mabi.201300134 URL |
| [15] |
Tang L, Wang Y, Gao S, et al. Biochemical characteristics and molecular mechanism of an exo-type alginate lyase VxAly7D and its use for the preparation of unsaturated monosaccharides[J]. Biotechnol Biofuels, 2020, 13:99.
doi: 10.1186/s13068-020-01738-4 URL |
| [16] |
Lu D, Zhang Q, Wang S, et al. Biochemical characteristics and synergistic effect of two novel alginate lyases from Photobacterium sp. FC615[J]. Biotechnol Biofuels, 2019, 12:260.
doi: 10.1186/s13068-019-1600-y URL |
| [17] |
Yamasaki M, Moriwaki S, Miyake O, et al. Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7:a novel alginate lyase with a beta-sandwich fold[J]. J Biol Chem, 2004, 279(30):31863-31872.
doi: 10.1074/jbc.M402466200 URL |
| [18] |
Kobayashi T, Uchimura K, Miyazaki M, et al. A new high-alkaline alginate lyase from a deep-sea bacterium Agarivorans sp[J]. Extremophiles, 2009, 13(1):121-129.
doi: 10.1007/s00792-008-0201-7 pmid: 19002649 |
| [19] |
Inoue A, Anraku M, Nakagawa S, et al. Discovery of a novel alginate lyase from Nitratiruptor sp. SB155-2 thriving at deep-sea hydrothermal vents and identification of the residues responsible for its heat stability[J]. J Biol Chem, 2016, 291(30):15551-15563.
doi: 10.1074/jbc.M115.713230 URL |
| [20] |
Li S, Wang L, Chen X, et al. Cloning, expression, and biochemical characterization of two new oligoalginate lyases with synergistic degradation capability[J]. Mar Biotechnol:NY, 2018, 20(1):75-86.
doi: 10.1007/s10126-017-9788-y URL |
| [1] | ZHAO Sai-sai, ZHANG Xiao-dan, JIA Xiao-yan, TAO Da-wei, LIU Ke-yu, NING Xi-bin. Investigation on the Complex Mutagenesis Selection of High-yield Nitrate Reductase Strain Staphylococcus simulans ZSJ6 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2023, 39(4): 103-113. |
| [2] | ZHANG Kai-ping, LIU Yan-li, TU Mian-liang, LI Ji-wei, WU Wen-biao. Optimization of Producing Cellulase by Aspergillus fumigatus A-16 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2022, 38(9): 215-225. |
| [3] | CAO Ru-fei, LI Ze-xuan, XU Huan, ZHANG Sha, ZHANG Min-min, DAI Feng, DUAN Xiao-lei. Expression,Purification,and Crystallization of Pif1 Helicase from Bacteroides fragilis [J]. Biotechnology Bulletin, 2021, 37(9): 180-190. |
| [4] | TIAN Jia-hui, FENG Jia-li, LU Jun-hua, MAO Lin-jing, HU Zhu-ran, WANG Ying, CHU Jie. Isolation,Purification and Characterization of Laccase LacT-1 from Cerrena unicolor [J]. Biotechnology Bulletin, 2021, 37(8): 186-194. |
| [5] | DUAN Xu-guo, ZHANG Yu-hua, HUANG Ting-ting, DING Qian, LUAN Shu-yue, ZHU Qiu-yu. Synergetic Enhancing the Soluble Expression of Thermotoga maritima α-Glucan Phosphorylase by Chemical Chaperones and Induction Condition Optimization [J]. Biotechnology Bulletin, 2021, 37(8): 233-242. |
| [6] | LIU Shan, YE Wei, ZHU Mu-zi, LI Sai-ni, DENG Zhang-shuang, ZHANG Wei-min. Cloning,Expression and Characterization of a Novel Acyltransferase GPAT [J]. Biotechnology Bulletin, 2021, 37(11): 257-266. |
| [7] | ZHAO Hai-yan, SONG Chen-bin, LIU Zheng-ya, MA Xing-rong, SHANG Hui-hui, LI An-hua, GUAN Xian-jun, WANG Jian-she. Cloning,Recombinant Expression and Enzymatic Properties of α-Amylase Gene from Laceyella sp. [J]. Biotechnology Bulletin, 2020, 36(8): 23-33. |
| [8] | ZHAO Zhen, WANG Sha-sha, LÜ Xing-xing, TAO Yan, XIE Jing, QIAN Yun-fang. Heterologous Expression of Cyclina sinensis Mytimacin Antibacterial Peptide Based on Recombinant Pichia pastoris [J]. Biotechnology Bulletin, 2020, 36(5): 150-158. |
| [9] | ZHU Cai-lin, LÜ Xiang, XIA Xiao-le. Effect of Site-directed Mutagenesis of Amino Acids in Lid Region on the Enzymatic Properties of T1 Lipase [J]. Biotechnology Bulletin, 2020, 36(11): 94-102. |
| [10] | GAO Yun-shan, LIU Dan-dan, XU Jun-lin, SANG Yu-nong, LIANG Xia-xia, LIU Jian-xin, WANG Wen-bin. Recombinant Expression and Immunogenicity Analysis of the Porin Protein OmpF of Aeromonas hydrophila [J]. Biotechnology Bulletin, 2019, 35(9): 234-243. |
| [11] | GUO Jing-jing, GUO Lei-lei, ZHAO Yun-xiu, DAI Yi-jun. Research on the NAMase of Ensifer meliloti 1021 and Regulation Mechanism of 3-Cyanopyridine [J]. Biotechnology Bulletin, 2019, 35(8): 51-58. |
| [12] | ZHANG Ya-li, TAO Yan, XIE Jing, QIAN Yun-fang. Recombinant Expression of Mytilus coruscus Mytilin-1 Mature Peptide in Pichia pastoris and Its Antibacterial Activity [J]. Biotechnology Bulletin, 2019, 35(7): 54-60. |
| [13] | ZHANG Qing-fang, PANG Fei, YU Shuang, XIAO Jing-hui, DOU Shao-hua, CHI Nai-yu. Screening and Identification of High Uricase-producting Strain from Marine and the Enzymatic Properties [J]. Biotechnology Bulletin, 2019, 35(7): 61-69. |
| [14] | XU Shan ,LI Ren-qiang ,ZHENG Zhen-hua ,ZHANG Yun ,SUN Ai-jun ,HU Yun-feng. Properties of Extracellular Protease of Microbe DH-2 from Mangrove and Optimization of Enzyme Producing Conditions [J]. Biotechnology Bulletin, 2018, 34(6): 120-127. |
| [15] | QIN Ri-tian, XIE Zhan-ling. Isolation,Purification,Characterization and Structural Analysis of a Pectinase PGL1 Produced by Fusarium sp. Q7-31T [J]. Biotechnology Bulletin, 2018, 34(4): 151-160. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||