








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (5): 94-102.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0967
Previous Articles Next Articles
SUN Bo1,2( ), WANG Rui1, HUO Yong-xue1, GE Jian-rong1, KUANG Meng2(
), WANG Rui1, HUO Yong-xue1, GE Jian-rong1, KUANG Meng2( ), WANG Feng-ge1(
), WANG Feng-ge1( )
)
Received:2023-10-18
Online:2024-05-26
Published:2024-03-28
Contact:
KUANG Meng, WANG Feng-ge
E-mail:sunbo990818@163.com;kuangmeng007@163.com;gege0106@163.com
SUN Bo, WANG Rui, HUO Yong-xue, GE Jian-rong, KUANG Meng, WANG Feng-ge. Design of Universal Tailed-sequence and Establishment of the Universal System Suitable for Multiple Species[J]. Biotechnology Bulletin, 2024, 40(5): 94-102.
| 反应组分 Reaction component | 荧光引物扩增体系 Fluorescent primer amplification system | TP-M13-SSR扩增体系 TP-M13-SSR amplification system | UTS通用扩增体系 UTS universal amplification system |
|---|---|---|---|
| ddH2O | / | / | / |
| 10×Buffer | 1× | 1× | 1× |
| MgCl2 | 2.5 mmol/L | 2.5 mmol/L | 2.5 mmol/L |
| dNTPs | 0.15 mmol/L each | 0.15 mmol/L each | 0.15 mmol/L each |
| Taq 酶 Taq enzyme | 1.0 U | 1.0 U | 1.0 U |
| 上游引物 Forward primer | 0.25 μmol/L | 0.015 μmol/L | 0.03 μmol/L |
| 下游引物 Reverse primer | 0.25 μmol/L | 0.1 μmol/L | 0.1 μmol/L |
| 通用尾巴序列 Universal tailed-sequence | / | 0.1 μmol/L | 0.1 μmol/L |
| 模板DNA Template DNA | 5 ng/μL | 5 ng/μL | 5 ng/μL |
Table 1 PCR amplification system of fluorescent primers and universal primer sequences
| 反应组分 Reaction component | 荧光引物扩增体系 Fluorescent primer amplification system | TP-M13-SSR扩增体系 TP-M13-SSR amplification system | UTS通用扩增体系 UTS universal amplification system |
|---|---|---|---|
| ddH2O | / | / | / |
| 10×Buffer | 1× | 1× | 1× |
| MgCl2 | 2.5 mmol/L | 2.5 mmol/L | 2.5 mmol/L |
| dNTPs | 0.15 mmol/L each | 0.15 mmol/L each | 0.15 mmol/L each |
| Taq 酶 Taq enzyme | 1.0 U | 1.0 U | 1.0 U |
| 上游引物 Forward primer | 0.25 μmol/L | 0.015 μmol/L | 0.03 μmol/L |
| 下游引物 Reverse primer | 0.25 μmol/L | 0.1 μmol/L | 0.1 μmol/L |
| 通用尾巴序列 Universal tailed-sequence | / | 0.1 μmol/L | 0.1 μmol/L |
| 模板DNA Template DNA | 5 ng/μL | 5 ng/μL | 5 ng/μL |
| 通用尾巴序列类别 UTS category | 扩增程序 Amplification program |
|---|---|
| UTS通用扩增程序 UTS universal amplification program | 95℃,5 min; 95℃,40 s,55℃,35 s,72℃,45 s,35 cycle; 72℃,10 min;4℃ store |
| TP-M13-SSR扩增程序 TP-M13-SSR amplification program | 95℃,5 min; 95℃,40 s,Primer annealing temperature,35 s,72℃,45 s,8 cycle; 95℃,40 s,M13 Tailed-sequence annealing temperature,35 s,72℃,45 s,30 cycle; 72℃,10 min;4℃ |
Table 2 PCR amplification program for UTS and M13
| 通用尾巴序列类别 UTS category | 扩增程序 Amplification program |
|---|---|
| UTS通用扩增程序 UTS universal amplification program | 95℃,5 min; 95℃,40 s,55℃,35 s,72℃,45 s,35 cycle; 72℃,10 min;4℃ store |
| TP-M13-SSR扩增程序 TP-M13-SSR amplification program | 95℃,5 min; 95℃,40 s,Primer annealing temperature,35 s,72℃,45 s,8 cycle; 95℃,40 s,M13 Tailed-sequence annealing temperature,35 s,72℃,45 s,30 cycle; 72℃,10 min;4℃ |
| UTS编号UTS code | 汉字字组 Chinese character | 编码后序列 Sequence after encoding(5'-3') | 序列GC含量 GC content of sequence/% | 发卡结构Tm Hairpin Tm value/℃ | 同源二聚体Tm值 Self-dimer Tm value/℃ |
|---|---|---|---|---|---|
| UTS01 | 孙擘(Sun Bo) | GACCAAGCACACGACAACTGACAT | 50.0 | / | -29.7 |
| UTS02 | 电泳(Dian Yong) | GACGACCTAGCCGACAAGTGAGTG | 58.3 | / | 9.3 |
| UTS03 | 荧光(Ying Guang) | GAATATGCAACGGACCATCCATAC | 45.8 | 35.9 | -25.8 |
| UTS04 | 基因(Ji Yin) | GACCACGGAGAAGACCACAGAATT | 50.0 | / | -19.6 |
| UTS05 | 分子(Fen Zi) | GACCATATATCAGACCAAGCACTT | 41.7 | / | -33.0 |
| UTS06 | 玉米(Yu Mi) | GACGATGAATACGACGAGTCAGTG | 50.0 | 34.7 | -0.4 |
Table 3 Evaluation of primers designed by UTS design tool
| UTS编号UTS code | 汉字字组 Chinese character | 编码后序列 Sequence after encoding(5'-3') | 序列GC含量 GC content of sequence/% | 发卡结构Tm Hairpin Tm value/℃ | 同源二聚体Tm值 Self-dimer Tm value/℃ |
|---|---|---|---|---|---|
| UTS01 | 孙擘(Sun Bo) | GACCAAGCACACGACAACTGACAT | 50.0 | / | -29.7 |
| UTS02 | 电泳(Dian Yong) | GACGACCTAGCCGACAAGTGAGTG | 58.3 | / | 9.3 |
| UTS03 | 荧光(Ying Guang) | GAATATGCAACGGACCATCCATAC | 45.8 | 35.9 | -25.8 |
| UTS04 | 基因(Ji Yin) | GACCACGGAGAAGACCACAGAATT | 50.0 | / | -19.6 |
| UTS05 | 分子(Fen Zi) | GACCATATATCAGACCAAGCACTT | 41.7 | / | -33.0 |
| UTS06 | 玉米(Yu Mi) | GACGATGAATACGACGAGTCAGTG | 50.0 | 34.7 | -0.4 |
| 物种名称Species name | 通用引物名称Universal primer name | ||||||
|---|---|---|---|---|---|---|---|
| M13 | UTS01 | UTS02 | UTS03 | UTS04 | UTS05 | UTS06 | |
| 玉米Zea mays L. | 0.004 | 8.2 | 2.1 | 0.13 | 0.52 | 0.52 | 0.13 |
| 水稻Oryza sativa L. | 0.026 | 19 | 4.7 | 4.7 | 4.7 | 4.7 | 1.2 |
| 小麦Triticum aestivum L. | 0.006 | 0.061 | 15 | 15 | 3.8 | 0.96 | 3.8 |
| 大豆Glycine max (L.)Merr. | 0.013 | 1.8 | 29 | 29 | 7.2 | 0.45 | 7.2 |
| 大麦Hordeum vulgare L. | 0.004 | 0.45 | 7 | 7 | 1.8 | 1.8 | 1.8 |
| 高粱Sorghum bicolor (L.)Moench | 3.7 | 8.7 | 34 | 2.2 | 2.2 | 8.7 | 2.2 |
| 甘薯Dioscorea esculenta (Lour.)Burkill | 1.7 | 10 | 2.6 | 0.67 | 0.67 | 2.6 | 0.17 |
| 马铃薯Solanum tuberosum L. | 0.022 | 3.2 | 50 | 3.2 | 0.21 | 0.81 | 50 |
| 棉花Gossypium hirsutum L. | 0.39 | 3.1 | 3.1 | 12 | 3.1 | 0.2 | 3.1 |
| 向日葵Helianthus annuus L. | 0.75 | 1.5 | 1.5 | 6 | 6 | 1.5 | 0.38 |
| 甘蔗Saccharum officinarum L. | 0.5 | 3.6 | 0.91 | 3.6 | 0.23 | 0.23 | 0.23 |
| 番茄Solanum lycopersicum L. | 0.024 | 21 | 5.2 | 1.3 | 5.2 | 1.3 | 21 |
| 黄瓜Cucumis sativus L. | 4.9 | 0.56 | 2.2 | 8.8 | 0.56 | 0.56 | 0.56 |
| 辣椒Capsicum annuum L. | 1.8 | 16 | 4.1 | 4.1 | 0.27 | 1 | 0.27 |
| 西瓜Citrullus lanatus subsp. Vuaris | 7.4 | 13 | 0.21 | 0.21 | 3.2 | 0.81 | 0.81 |
| 甜瓜Cucumis melo L. | 2 | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 |
| 梨Pyrus L. | 5.9 | 0.065 | 16 | 0.065 | 0.065 | 1 | 4 |
| 枣Ziziphus jujuba(L.)Lam. | 6.7 | 0.22 | 3.4 | 3.4 | 0.22 | 0.86 | 13 |
| 菜豆Phaseolus vulgaris L. | 0.26 | 0.47 | 7.3 | 1.8 | 1.8 | 0.47 | 0.12 |
| 苹果Malus pumila Mill. | 1.6 | 0.15 | 2.3 | 0.58 | 2.3 | 0.58 | 0.15 |
Table 4 BLAST assessments for M13 universal primers and six UTS sequences across the genomes of 20 crops
| 物种名称Species name | 通用引物名称Universal primer name | ||||||
|---|---|---|---|---|---|---|---|
| M13 | UTS01 | UTS02 | UTS03 | UTS04 | UTS05 | UTS06 | |
| 玉米Zea mays L. | 0.004 | 8.2 | 2.1 | 0.13 | 0.52 | 0.52 | 0.13 |
| 水稻Oryza sativa L. | 0.026 | 19 | 4.7 | 4.7 | 4.7 | 4.7 | 1.2 |
| 小麦Triticum aestivum L. | 0.006 | 0.061 | 15 | 15 | 3.8 | 0.96 | 3.8 |
| 大豆Glycine max (L.)Merr. | 0.013 | 1.8 | 29 | 29 | 7.2 | 0.45 | 7.2 |
| 大麦Hordeum vulgare L. | 0.004 | 0.45 | 7 | 7 | 1.8 | 1.8 | 1.8 |
| 高粱Sorghum bicolor (L.)Moench | 3.7 | 8.7 | 34 | 2.2 | 2.2 | 8.7 | 2.2 |
| 甘薯Dioscorea esculenta (Lour.)Burkill | 1.7 | 10 | 2.6 | 0.67 | 0.67 | 2.6 | 0.17 |
| 马铃薯Solanum tuberosum L. | 0.022 | 3.2 | 50 | 3.2 | 0.21 | 0.81 | 50 |
| 棉花Gossypium hirsutum L. | 0.39 | 3.1 | 3.1 | 12 | 3.1 | 0.2 | 3.1 |
| 向日葵Helianthus annuus L. | 0.75 | 1.5 | 1.5 | 6 | 6 | 1.5 | 0.38 |
| 甘蔗Saccharum officinarum L. | 0.5 | 3.6 | 0.91 | 3.6 | 0.23 | 0.23 | 0.23 |
| 番茄Solanum lycopersicum L. | 0.024 | 21 | 5.2 | 1.3 | 5.2 | 1.3 | 21 |
| 黄瓜Cucumis sativus L. | 4.9 | 0.56 | 2.2 | 8.8 | 0.56 | 0.56 | 0.56 |
| 辣椒Capsicum annuum L. | 1.8 | 16 | 4.1 | 4.1 | 0.27 | 1 | 0.27 |
| 西瓜Citrullus lanatus subsp. Vuaris | 7.4 | 13 | 0.21 | 0.21 | 3.2 | 0.81 | 0.81 |
| 甜瓜Cucumis melo L. | 2 | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 |
| 梨Pyrus L. | 5.9 | 0.065 | 16 | 0.065 | 0.065 | 1 | 4 |
| 枣Ziziphus jujuba(L.)Lam. | 6.7 | 0.22 | 3.4 | 3.4 | 0.22 | 0.86 | 13 |
| 菜豆Phaseolus vulgaris L. | 0.26 | 0.47 | 7.3 | 1.8 | 1.8 | 0.47 | 0.12 |
| 苹果Malus pumila Mill. | 1.6 | 0.15 | 2.3 | 0.58 | 2.3 | 0.58 | 0.15 |
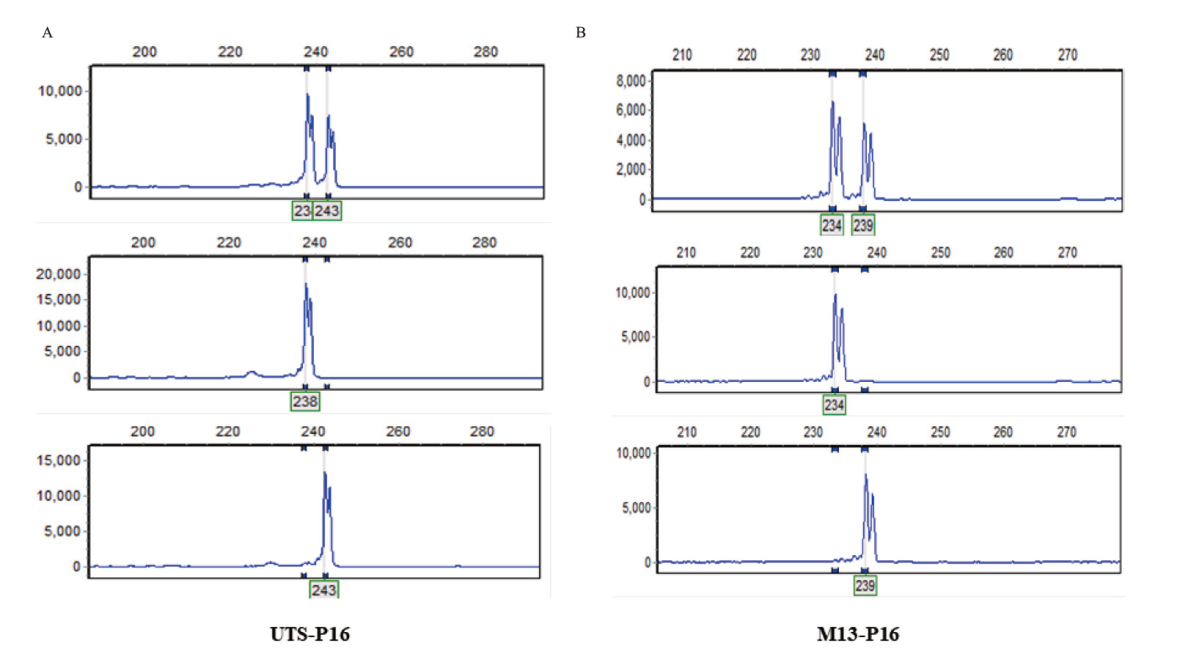
Fig. 1 Comparison of amplified peaks between UTS and M13 tailed-sequence reaction procedures and reaction system A: Amplification results of UTS01-P16 on three corn samples. B: Amplification results of M13-P16 on three corn samples
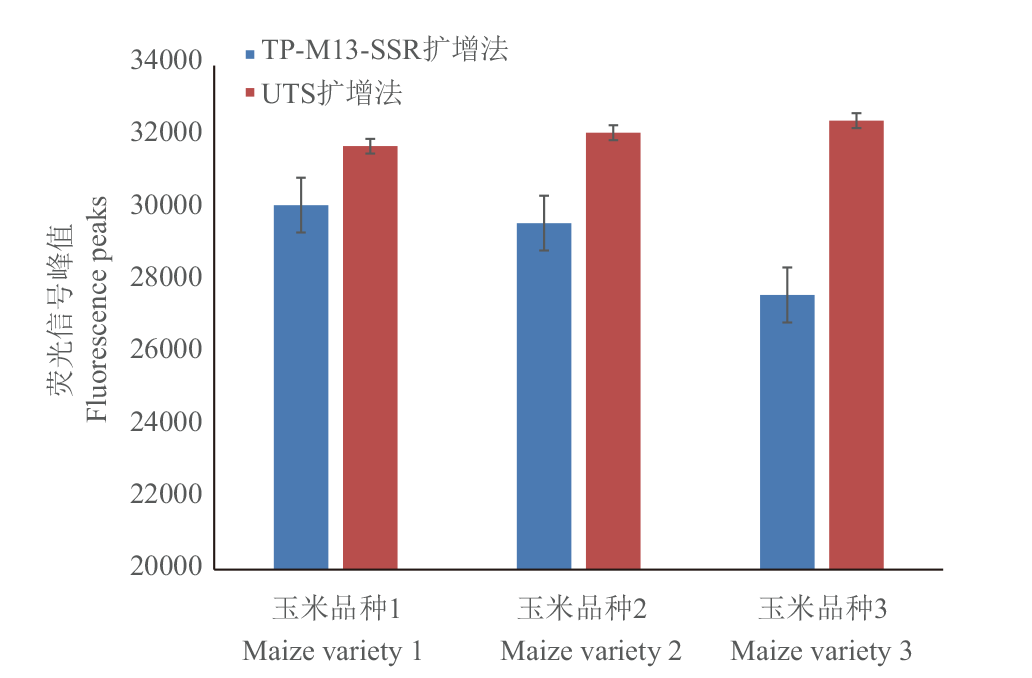
Fig. 2 Comparison of amplification peaks between UTS universal primers and M13 reaction procedures and reaction systems Error line in the figure refers to the standard deviation
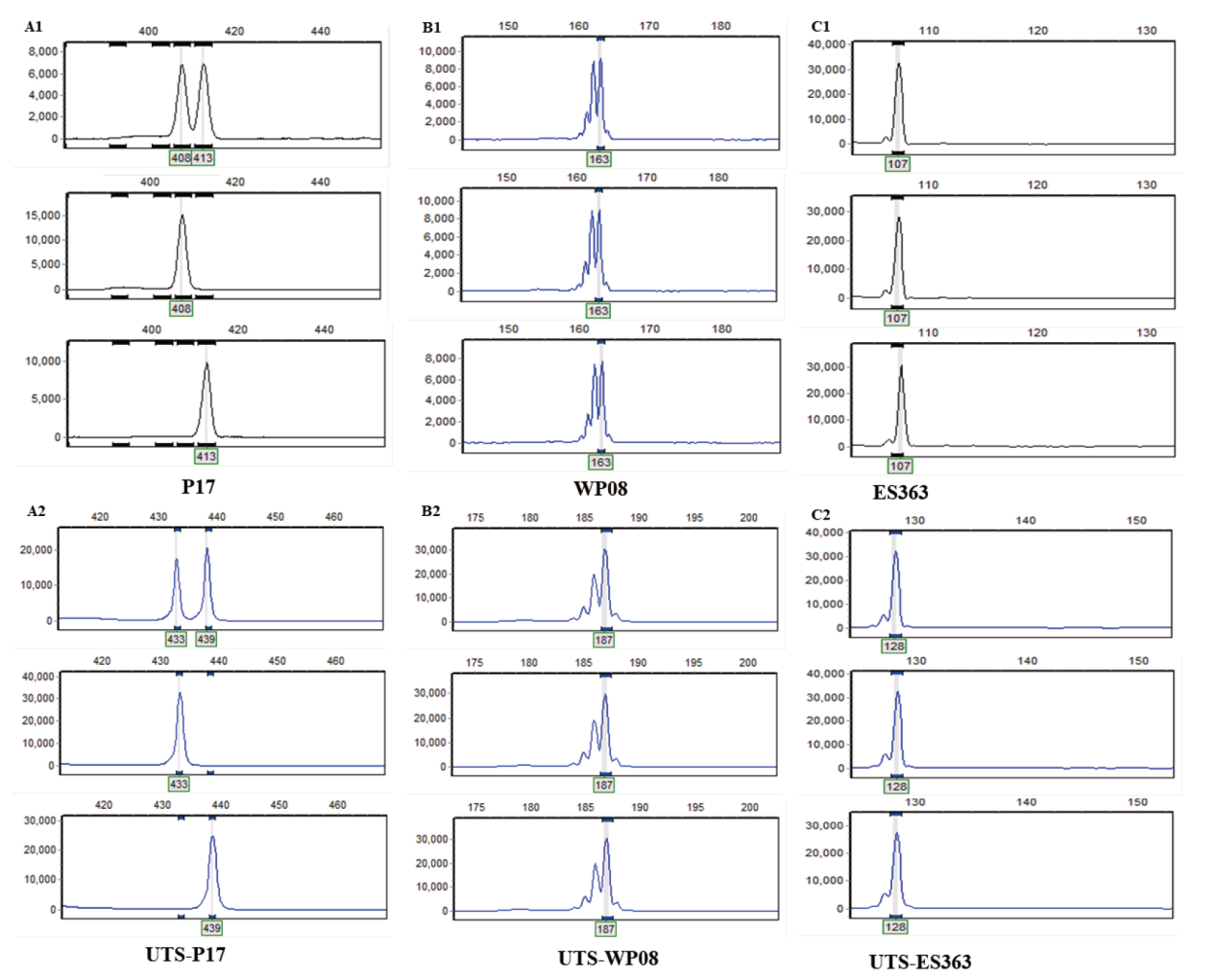
Fig. 3 Fluorescent capillary electropherograms of different crops using fluorescent primers and UTS A1: Amplified results of UTS01-P17 on three corn samples. A2: Amplified results of P17 fluorescent primer on three corn germplasm resources. B1: Amplified results of UTS01-WP08 on three watermelon germplasm resources. B2: Amplified results of WP08 fluorescent primer on three watermelon germplasm resources. C1: Amplified results of UTS01-ES36308 on three pepper germplasm resources. C2: Amplified results of ES363 fluorescent primer on three watermelon germplasm resources
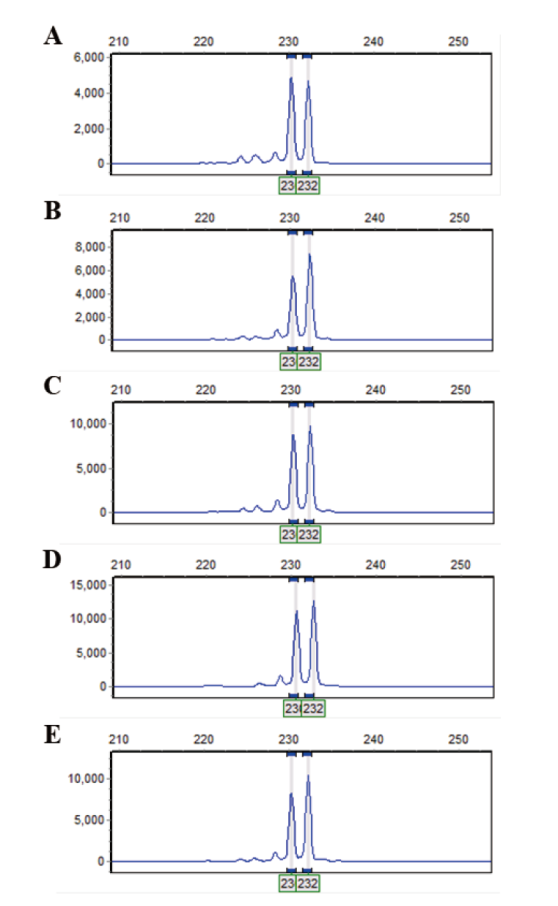
Fig. 4 Amplified results of UTS02-UTS06 ligated to tomato SOL_SSR145 primer A: Amplified results of UTS02- SOL_SSR145. B: Amplified results of UTS03- SOL_SSR145. C: Amplified results of UTS04- SOL_SSR145. D: Amplified results of UTS05- SOL_SSR145. E: Amplified results of UTS06- SOL_SSR145
| 物种名称 Species | 测试引物总数Total number of primers tested | M13扩增成功率M13 amplified success rate/% | 单UTS扩增成功率Single UTS amplified success rate/% | 两个UTS扩增成功率Two UTS amplified success rate/% |
|---|---|---|---|---|
| 玉米Z. mays | 40 | 72.5 | 85.0 | 95.0 |
| 西瓜C. lanatus | 28 | 96.4 | 100.0 | 100.0 |
| 番茄S. lycopersicum | 24 | 87.5 | 91.7 | 100.0 |
| 辣椒C. annuum | 22 | 86.4 | 86.4 | 95.5 |
Table 5 UTS- and M13-amplified effects on various crops
| 物种名称 Species | 测试引物总数Total number of primers tested | M13扩增成功率M13 amplified success rate/% | 单UTS扩增成功率Single UTS amplified success rate/% | 两个UTS扩增成功率Two UTS amplified success rate/% |
|---|---|---|---|---|
| 玉米Z. mays | 40 | 72.5 | 85.0 | 95.0 |
| 西瓜C. lanatus | 28 | 96.4 | 100.0 | 100.0 |
| 番茄S. lycopersicum | 24 | 87.5 | 91.7 | 100.0 |
| 辣椒C. annuum | 22 | 86.4 | 86.4 | 95.5 |
| [1] | 谭智广. 低功耗聚合酶链式反应温度控制系统的研制[D]. 大连: 大连理工大学, 2023. |
| Tan GZ. Development of low power consumption temperature control system of PCR[D]. Dalian: Dalian University of Technology, 2023. | |
| [2] | Zhu HL, Zhang HQ, Xu Y, et al. PCR past, present and future[J]. BioTechniques, 2020, 69(4): 317-325. |
| [3] | 郑戈文. 农作物种子检测中常用电泳方法的比较分析[J]. 中国种业, 2018(11): 35-37. |
| Zheng GW. Comparative analysis of electrophoresis methods commonly used in crop seed detection[J]. China Seed Ind, 2018(11): 35-37. | |
| [4] | 王爱娜, 王灏, 李殿荣, 等. 琼脂糖与聚丙烯酰胺凝胶电泳在SSR鉴定杂交油菜种子纯度中的比较[J]. 西北农业学报, 2012, 21(8): 101-106. |
| Wang AN, Wang H, Li DR, et al. Comparison of agarose and polyacrylamide gel electrophoresis for SSR in purity identification of qinyou 33 hybrid rape seed[J]. Acta Agric Boreali Occidentalis Sin, 2012, 21(8): 101-106. | |
| [5] | 王竹林, 杨睿, 刘联正, 等. 聚丙烯酰胺凝胶电泳银染的影响因素[J]. 实验室研究与探索, 2011, 30(9): 15-17, 113. |
| Wang ZL, Yang R, Liu LZ, et al. Factors of silver staining of polyacrylamide gel electrophoresis[J]. Res Explor Lab, 2011, 30(9): 15-17, 113. | |
| [6] |
冯琬淇, 张福顺, 刘乃新. 毛细管与聚丙烯酰胺凝胶电泳检测甜菜SSR位点的比较研究[J]. 中国农学通报, 2022, 38(36): 34-41.
doi: 10.11924/j.issn.1000-6850.casb2022-0412 |
|
Feng WQ, Zhang FS, Liu NX. SSR polymorphism in sugar beet: detection by polyacrylamide gel electrophoresis and capillary electrophoresis[J]. Chin Agric Sci Bull, 2022, 38(36): 34-41.
doi: 10.11924/j.issn.1000-6850.casb2022-0412 |
|
| [7] | 刘文彬, 许理文, 王凤格, 等. 基于两种荧光毛细管电泳平台筛选评估玉米新型SSR引物[J]. 玉米科学, 2017, 25(2): 24-30. |
| Liu WB, Xu LW, Wang FG, et al. Evaluating and screening new maize SSR primer based on two kinds of fluorescent capillary eelectrophoresis platform[J]. J Maize Sci, 2017, 25(2): 24-30. | |
| [8] |
管俊娇, 杨晓洪, 张鹏, 等. 基于荧光检测技术的青稞品种鉴定方法的建立[J]. 生物技术通报, 2021, 37(11): 293-302.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-1494 |
| Guan JJ, Yang XH, Zhang P, et al. Establishment of a method identifying the varieties of hulless barley based on fluorescence detection technology[J]. Biotechnol Bull, 2021, 37(11): 293-302. | |
| [9] | 方梅, 胡波, 侯媛媛, 等. 毛细管电泳在基因研究中的应用[J]. 生物技术通报, 2006(2): 60-66. |
| Fang M, Hu B, Hou YY, et al. The application of capillary electrophoresis to gene analysis[J]. Biotechnol Bull, 2006(2): 60-66. | |
| [10] | 李会勇, 王天宇, 黎裕, 等. TP-M13自动荧光检测法在高粱SSR基因型鉴定中的应用[J]. 植物遗传资源学报, 2005, 6(1): 68-70. |
| Li HY, Wang TY, Li Y, et al. Application of the TP-M13 automated fluorescent-lablled system of SSR genotyping in sorghum[J]. J Plant Genet Resour, 2005, 6(1): 68-70. | |
| [11] |
Schuelke M. An economic method for the fluorescent labeling of PCR fragments[J]. Nat Biotechnol, 2000, 18(2): 233-234.
doi: 10.1038/72708 pmid: 10657137 |
| [12] | 匡猛, 王凤格, 赵久然, 等. 玉米SSR引物尾巴序列的设计方法[J]. 农业生物技术学报, 2007, 15(6): 1072-1073. |
| Kuang M, Wang FG, Zhao JR, et al. A method of designing tail sequence for maize SSR primers[J]. J Agric Biotechnol, 2007, 15(6): 1072-1073. | |
| [13] |
曾斌, 马鑫鑫, 余镇藩, 等. 新疆野扁桃的TP-M13-SSR荧光标记引物的筛选[J]. 中国农学通报, 2019, 35(1): 45-49.
doi: 10.11924/j.issn.1000-6850.casb18070067 |
| Zeng B, Ma XX, Yu ZF, et al. Wild almond(Prunus tenella batsch.) in Xinjiang: screening of TP-M13-SSR primers in capillary fluorescent electrophoresis[J]. Chin Agric Sci Bull, 2019, 35(1): 45-49. | |
| [14] | 赵紫薇, 任洁, 刘亚维, 等. 一种适用于玉米胚乳DNA提取的新方法[J]. 分子植物育种, 2023: 1-10. |
| Zhao ZW, Ren J, Liu YW, et al. A New Method for DNA Extraction from Maize Endosperm[J]. Molecular Plant Breeding, 2023: 1-10. | |
| [15] | 国家市场监督管理总局, 国家标准化管理委员会. 主要农作物品种真实性和纯度SSR分子标记检测玉米: GB/T 39914—2021[S]. 北京: 中国标准出版社, 2021. |
| State Administration of Market Supervision and administration, Standardization Administration of the People's Republic of China. Variety genuineness and purity testing of main crops with SSR markers—Maize: GB/T 39914—2021[S]. Beijing: Standards Press of China, 2021. | |
| [16] | 中华人民共和国农业部. 番茄品种鉴定技术规程 Indel分子标记法: NY/T 2471—2013[S]. 北京: 中国农业出版社, 2014. |
| Ministry of Agriculture of the People's Republic of China. Identification of tomato varieties—Indel marker method: NY/T 2471—2013[S]. Beijing: China Agriculture Press, 2014. | |
| [17] | 中华人民共和国农业部. 西瓜品种鉴定技术规程 SSR分子标记法: NY/T 2472—2013[S]. 北京: 中国农业出版社, 2014. |
| Ministry of Agriculture of the People's Republic of China. Identification of watermelon varieties—SSR marker method: NY/T 2472—2013[S]. Beijing: China Agriculture Press, 2014. | |
| [18] | 中华人民共和国农业部. 辣椒品种鉴定技术规程 SSR分子标记法: NY/T 2475—2013[S]. 北京: 中国农业出版社, 2014. |
| Ministry of Agriculture of the People's Republic of China. Identification of pepper varieties—SSR marker method: NY/T 2475—2013[S]. Beijing: China Agriculture Press, 2014. | |
| [19] | 刘志斋, 王天宇, 黎裕. TP-M13-SSR技术及其在玉米遗传多样性研究中的应用[J]. 玉米科学, 2007, 15(6): 10-15, 31. |
| Liu ZZ, Wang TY, Li Y. TP-M13-SSR technique and its applications in the analysis of genetic diversity in maize[J]. J Maize Sci, 2007, 15(6): 10-15, 31. | |
| [20] | 李小娟, 刘文雅, 何彦峰, 等. 葫芦藓叶绿体基因组密码子使用模式分析[J]. 分子植物育种, 2023: 1-15. |
| Li XJ, Liu WY, He YF, et al. Analysis of codon usage patterns in chloroplast genomes of funaria hygrometrica[J]. Molecular Plant Breeding, 2023: 1-15. | |
| [21] |
仇学文, 李丹, 甘玉迪, 等. 豇豆叶绿体基因组密码子使用偏好性分析[J]. 核农学报, 2023, 37(6): 1118-1129.
doi: 10.11869/j.issn.1000-8551.2023.06.1118 |
|
Qiu XW, Li D, Gan YD, et al. Analysis of codon usage bias in the cowpea chloroplast genome[J]. J Nucl Agric Sci, 2023, 37(6): 1118-1129.
doi: 10.11869/j.issn.1000-8551.2023.06.1118 |
|
| [22] | 雷佳欣, 张丽娟, 高丹丹, 等. 文冠果基因组密码子偏好性分析[J]. 西北林学院学报, 2023, 38(4): 104-110. |
| Lei JX, Zhang LJ, Gao DD, et al. Codon usage bias of Xanthoceras sorbifolium genome[J]. J Northwest For Univ, 2023, 38(4): 104-110. |
| [1] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [2] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [3] | ZHENG Qiao, LIN Hua, XU Hao, AN Wei, XUE Chang-hua, ZHANG Jing, HAN Guo-quan. Establishment and Application of Multiplex Droplet Digital PCR for SARS-CoV-2 Omicron Variant [J]. Biotechnology Bulletin, 2024, 40(2): 80-89. |
| [4] | YU Hui, WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun. Isolation and Functional Verification of Genes Responding to Iron and Cadmium Stresses in Lycium barbarum [J]. Biotechnology Bulletin, 2023, 39(7): 195-205. |
| [5] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [6] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [7] | SONG Hai-na, WU Xin-tong, YANG Lu-yu, GENG Xi-ning, ZHANG Hua-min, SONG Xiao-long. Selection and Validation of Reference Genes for RT-qPCR in Allium tuberosum Infected by Botrytis squamosa [J]. Biotechnology Bulletin, 2023, 39(3): 101-115. |
| [8] | LI Tian-shun, LI Chen-wei, WANG Jia, ZHU Long-Jiao, XU Wen-tao. Efficient Generation of Secondary Libraries During Functional Nucleic Acids Screening [J]. Biotechnology Bulletin, 2023, 39(3): 116-122. |
| [9] | YU Shi-zhou, CAO Ling-gai, WANG Shi-ze, LIU Yong, BIAN Wen-jie, REN Xue-liang. Development Core SNP Markers for Tobacco Germplasm Genotyping [J]. Biotechnology Bulletin, 2023, 39(3): 89-100. |
| [10] | MU De-tian, WAN Ling-yun, ZHANG Yao, WEI Shu-gen, LU Ying, FU Jin-e, TIAN Yi, PAN Li-mei, TANG Qi. House-keeping Genes Screening and Expression Patterns Analysis of Genes Involved in Alkaloid Biosynthesis in Uncaria rhynchophylla [J]. Biotechnology Bulletin, 2023, 39(2): 126-138. |
| [11] | TENG Meng-xin, XU Ya, HE Jing, WANG Qi, QIAO Fei, LI Jing-yang, LI Xin-guo. Cloning and Prokaryotic Expression Analysis of MaMC6 in Banana [J]. Biotechnology Bulletin, 2023, 39(12): 179-186. |
| [12] | LI Hui-jie, DONG Lian-hua, CHEN Gui-fang, LIU Si-yuan, YANG Jia-yi, YANG Jing-ya. Establishment of Droplet Digital PCR Assay for Quantitative Detection of Pseudomonas cocovenenans in Foods [J]. Biotechnology Bulletin, 2023, 39(1): 127-136. |
| [13] | HU Xue-ying, ZHANG Yue, GUO Ya-jie, QIU Tian-lei, GAO Min, SUN Xing-bin, WANG Xu-ming. Comparison in Antibiotic Resistance Genes Carried by Bacteriophages and Bacteria in Farmland Soil Amended with Different Fertilizers [J]. Biotechnology Bulletin, 2022, 38(9): 116-126. |
| [14] | CHENG Shen-wei, ZHANG Ke-qiang, LIANG Jun-feng, LIU Fu-yuan, GAO Xing-liang, DU Lian-zhu. Establishment of a Triple Droplet Digital PCR Quantitative Detection Method for Typical Pathogenic Bacteria in Livestock and Poultry Manure [J]. Biotechnology Bulletin, 2022, 38(9): 271-280. |
| [15] | CAO Ying-fang, ZHAO Xin, LIU Shuang, LI Rui-huan, LIU Na, XU Shi-yong, GAO Fang-rui, MA Hui, LAN Qing-kuo, TAN Jian-xin, WANG Yong. Establishment of Real-time Fluorescent Quantitative PCR Detection Method for Genetically Modified Herbicide-tolerant Soybean GE-J12 [J]. Biotechnology Bulletin, 2022, 38(7): 146-152. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||