








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (4): 255-263.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1074
Previous Articles Next Articles
ZHONG Yun1( ), LIN Chun1,2, LIU Zheng-jie1,2, DONG Chen-wen-hua1,2, MAO Zi-chao1,2(
), LIN Chun1,2, LIU Zheng-jie1,2, DONG Chen-wen-hua1,2, MAO Zi-chao1,2( ), LI Xing-yu1,2,3(
), LI Xing-yu1,2,3( )
)
Received:2023-11-14
Online:2024-04-26
Published:2024-04-30
Contact:
MAO Zi-chao, LI Xing-yu
E-mail:1292726737@qq.com;zmao@ynau.edu.cn;lixingyu@ynau.edu.cn
ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes[J]. Biotechnology Bulletin, 2024, 40(4): 255-263.
| 引物Primer | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| UAoSGT1 | F:TCTTGATGCGATTACAGCGG R:GAGATGGCTACAAGGTGAAAGG | 基因克隆 Gene clone |
| PGEX-AoSGT1 | F:GTGGATCCCC GAATTCCATGAGGAGCGGAGATTTCGAAG R:GGCCGCTCGA GTCGACCTACAAGGTGAAAGGAAGACAACACC | 原核表达 Prokaryotic expression |
| qRT-AoSGT1 | F:ATCCTTTGGGTGGTGATCCG R:AGAGCTTCAGCAACATGCGT | 实时荧光定量PCR RT-qPCR |
| EF1A | F:CTGGCCAGGGTGGTTCATGAT R:TAAGTCTGTTGAGATGCACC | 内参基因 Reference gene |
Table 1 PCR primer sequences used in this study
| 引物Primer | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| UAoSGT1 | F:TCTTGATGCGATTACAGCGG R:GAGATGGCTACAAGGTGAAAGG | 基因克隆 Gene clone |
| PGEX-AoSGT1 | F:GTGGATCCCC GAATTCCATGAGGAGCGGAGATTTCGAAG R:GGCCGCTCGA GTCGACCTACAAGGTGAAAGGAAGACAACACC | 原核表达 Prokaryotic expression |
| qRT-AoSGT1 | F:ATCCTTTGGGTGGTGATCCG R:AGAGCTTCAGCAACATGCGT | 实时荧光定量PCR RT-qPCR |
| EF1A | F:CTGGCCAGGGTGGTTCATGAT R:TAAGTCTGTTGAGATGCACC | 内参基因 Reference gene |
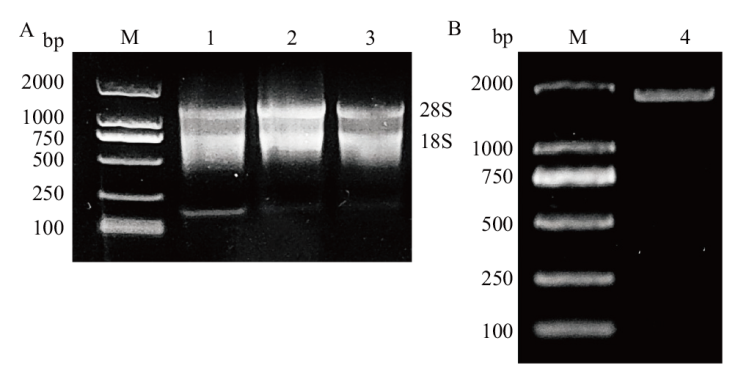
Fig. 1 Agarose gel electrophoresis of asparagus RNA and AOSGT1 gene amplification products A: Agarose gel electrophoresis of total RNA from various tissues of asparagus. B: Amplification of the AoSGT1 gene. M: DNA marker 2000. 1: Root. 2: Stem. 3: Flower. 4: AoSGT1
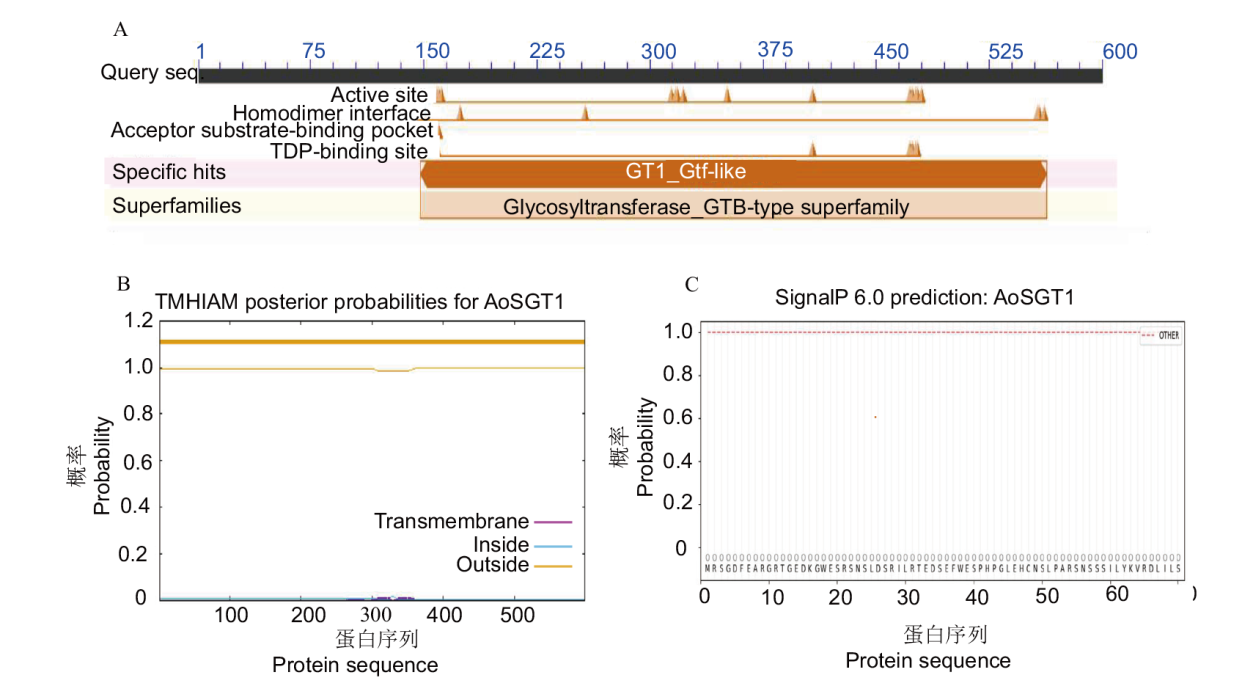
Fig. 2 Bioinformatics analysis of AoSGT1 A: NCBI conservative structural domain prediction. B: Transmembrane domain prediction. C: Signal peptide analysis
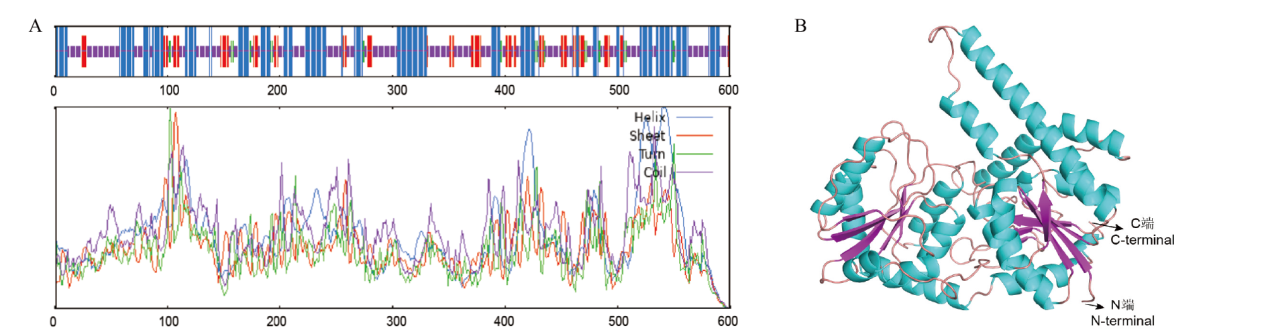
Fig. 3 Structural characteristics of asparagus AoSGT1 protein A: AoSGT1 secondary structure(Blue: α- Spiral; green: β- Corner; orange: irregular curl; purple: no secondary structure). B: AoSGT1 3D structure prediction(Blue: helix; pink: sheet; orange: loop)
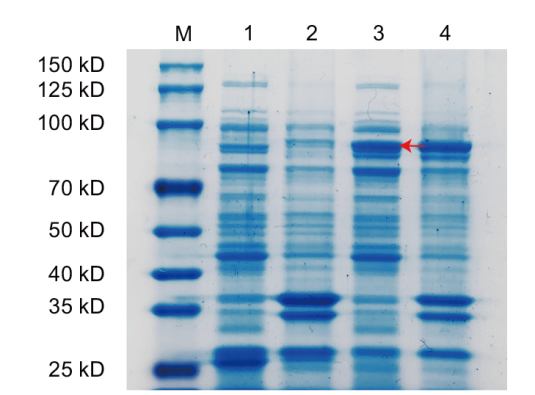
Fig. 7 SDS-PAGE gel electrophoresis of prokaryotic expression of AoSGT1 M: Protein marker. 1: Empty supernatant. 2: Empty sediment. 3: AoSGT1 supernatant. 4: AoSGT1 sediment
| [1] | 皇秋秋, 杜加欢, 尹俊玉, 等. 芦笋甾体皂苷合成与调控的研究进展[J]. 植物生理学报, 2020, 56(7): 1395-1407. |
| Huang QQ, Du JH, Yin JY, et al. Advances in biosynthesis and regulation of saponins from Asparagus officinalis[J]. Plant Physiol J, 2020, 56(7): 1395-1407. | |
| [2] |
Yu Q, Fan LP. Improving the bioactive ingredients and functions of asparagus from efficient to emerging processing technologies: a review[J]. Food Chem, 2021, 358: 129903.
doi: 10.1016/j.foodchem.2021.129903 URL |
| [3] |
Zhang HX, Birch J, Pei JJ, et al. Phytochemical compounds and biological activity in Asparagus roots: a review[J]. Int J Food Sci Tech, 2019, 54(4): 966-977.
doi: 10.1111/ijfs.2019.54.issue-4 URL |
| [4] | Li Y, Yang H, Li ZH, et al. Advances in the biosynthesis and molecular evolution of steroidal saponins in plants[J]. Int J Mol Sci, 2023, 24(3): 2620. |
| [5] |
Moreau RA, Nyström L, Whitaker BD, et al. Phytosterols and their derivatives: structural diversity, distribution, metabolism, analysis, and health-promoting uses[J]. Prog Lipid Res, 2018, 70: 35-61.
doi: S0163-7827(17)30062-0 pmid: 29627611 |
| [6] | Anwar Z, Hussain F. Steroidal saponins: an overview of medicinal uses[J]. Int J Chem & Biochem Sci, 2017,11: 20-24. |
| [7] |
Zhou C, Yang YH, Tian JY, et al. 22R- but not 22S-hydroxycholesterol is recruited for diosgenin biosynthesis[J]. Plant J, 2022, 109(4): 940-951.
doi: 10.1111/tpj.v109.4 URL |
| [8] |
Mohammadi M, Mashayekh T, Rashidi-Monfared S, et al. New insights into diosgenin biosynthesis pathway and its regulation in Trigonella foenum-graecum L[J]. Phytochem Anal, 2020, 31(2): 229-241.
doi: 10.1002/pca.v31.2 URL |
| [9] |
Yin Y, Gao LH, Zhang XN, et al. A cytochrome P450 monooxygenase responsible for the C-22 hydroxylation step in the Paris polyphylla steroidal saponin biosynthesis pathway[J]. Phytochemistry, 2018, 156: 116-123.
doi: 10.1016/j.phytochem.2018.09.005 URL |
| [10] | 秦晶晶, 孙春玉, 张美萍, 等. 植物UDP-糖基转移酶分类、功能以及进化[J]. 基因组学与应用生物学, 2018, 37(1): 440-450. |
| Qin JJ, Sun CY, Zhang MP, et al. Classification, function and evolution of plant UDP-glycosyltransferase[J]. Genom Appl Biol, 2018, 37(1): 440-450. | |
| [11] | Cantarel BL, Coutinho PM, Rancurel C, et al. The Carbohydrate-Active EnZymes database(CAZy): an expert resource for glycogenomics[J]. Nucleic Acids Res, 2009, 37(Database issue): D233-D238. |
| [12] |
Stucky DF, Arpin JC, Schrick K. Functional diversification of two UGT80 enzymes required for steryl glucoside synthesis in Arabidopsis[J]. J Exp Bot, 2015, 66(1): 189-201.
doi: 10.1093/jxb/eru410 URL |
| [13] | Tiwari P, Sangwan RS, Asha, et al. Molecular cloning and biochemical characterization of a recombinant sterol 3-O-glucosyltransferase from Gymnema sylvestre R.Br. catalyzing biosynthesis of steryl glucosides[J]. Biomed Res Int, 2014, 2014: 934351. |
| [14] |
Gao JH, Xu YH, Hua CK, et al. Molecular cloning and functional characterization of a sterol 3- O-glucosyltransferase involved in biosynthesis of steroidal saponins in Trigonella foenum-graecum[J]. Front Plant Sci, 2021, 12: 809579.
doi: 10.3389/fpls.2021.809579 URL |
| [15] | Li J, Liang Q, Li CF, et al. Comparative transcriptome analysis identifies putative genes involved in dioscin biosynthesis in Dioscorea zingiberensis[J]. Molecules, 2018, 23(2): 454. |
| [16] |
Song W, Zhang CC, Wu JL, et al. Characterization of three Paris polyphylla glycosyltransferases from different UGT families for steroid functionalization[J]. ACS Synth Biol, 2022, 11(4): 1669-1680.
doi: 10.1021/acssynbio.2c00103 pmid: 35286065 |
| [17] | Harkess A, Zhou JS, Xu CY, et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome[J]. Nat Commun, 2017, 8(1): 1279. |
| [18] | Cheng Q, Zeng LQ, Wen H, et al. Steroidal saponin profiles and their key genes for synthesis and regulation in Asparagus officinalis L. by joint analysis of metabolomics and transcriptomics[J]. BMC Plant Biol, 2023, 23(1): 207. |
| [19] |
Chen LQ, Zhang Y, Feng Y. Structural dissection of sterol glycosyltransferase UGT51 from Saccharomyces cerevisiae for substrate specificity[J]. J Struct Biol, 2018, 204(3): 371-379.
doi: 10.1016/j.jsb.2018.11.001 URL |
| [20] |
Upadhyay S, Jeena GS, et al. Shikha, Recent advances in steroidal saponins biosynthesis and in vitro production[J]. Planta, 2018, 248(3): 519-544.
doi: 10.1007/s00425-018-2911-0 pmid: 29748819 |
| [21] |
Liu M, Kong JQ. The enzymatic biosynthesis of acylated steroidal glycosides and their cytotoxic activity[J]. Acta Pharm Sin B, 2018, 8(6): 981-994.
doi: 10.1016/j.apsb.2018.04.006 pmid: 30505666 |
| [22] |
Zhang M, Li FD, Li K, et al. Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra[J]. J Am Chem Soc, 2020, 142(7): 3506-3512.
doi: 10.1021/jacs.9b12211 pmid: 31986016 |
| [23] |
Yang M, Fehl C, Lees KV, et al. Functional and informatics analysis enables glycosyltransferase activity prediction[J]. Nat Chem Biol, 2018, 14(12): 1109-1117.
doi: 10.1038/s41589-018-0154-9 pmid: 30420693 |
| [24] | Zhao XY, Zheng L, Si JJ, et al. Immunocytochemical localization of saikosaponin-d in vegetative organs of Bupleurum scorzonerifolium Willd[J]. Bot Stud, 2013, 54(1): 32. |
| [25] | Ye T, Song W, Zhang JJ, et al. Identification and functional characterization of DzS3GT, a cytoplasmic glycosyltransferase catalyzing biosynthesis of diosgenin 3-O-glucoside in Dioscorea zingiberensis[J]. Plant Cell Tissue Organ Cult PCTOC, 2017, 129(3): 399-410. |
| [26] |
Jaiswal Y, Liang ZT, Ho A, et al. A comparative tissue-specific metabolite analysis and determination of protodioscin content in Asparagus species used in traditional Chinese medicine and Ayurveda by use of laser microdissection, UHPLC-QTOF/MS and LC-MS/MS[J]. Phytochem Anal, 2014, 25(6): 514-528.
doi: 10.1002/pca.2522 URL |
| [27] | Yi TG, Yeoung YR, Choi IY, et al. Transcriptome analysis of Asparagus officinalis reveals genes involved in the biosynthesis of rutin and protodioscin[J]. PLoS One, 2019, 14(7): e0219973. |
| [28] |
Kohara A, Nakajima C, Hashimoto K, et al. A novel glucosyltransferase involved in steroid saponin biosynthesis in Solanum aculeatissimum[J]. Plant Mol Biol, 2005, 57(2): 225-239.
doi: 10.1007/s11103-004-7204-2 URL |
| [29] | 张磊, 唐永凯, 李红霞, 等. 促进原核表达蛋白可溶性的研究进展[J]. 中国生物工程杂志, 2021, 41(S1): 138-149. |
| Zhang L, Tang YK, Li HX, et al. Advances in promoting solubility of prokaryotic expressed proteins[J]. China Biotechnol, 2021, 41(S1): 138-149. | |
| [30] |
Hayat SMG, Farahani N, Golichenari B, et al. Recombinant protein expression in Escherichia coli(E. coli): what we need to know[J]. Curr Pharm Des, 2018, 24(6): 718-725.
doi: 10.2174/1381612824666180131121940 URL |
| [1] | DU Ze-guang, REN Shao-wen, ZHANG Feng-qin, LI Mei-lan, LI Gai-zhen, QI Xian-hui. Cloning,Expression and Functional Identification of BrMLP328 Gene in Brassica rapa subsp. pekinensis [J]. Biotechnology Bulletin, 2024, 40(4): 122-129. |
| [2] | GUO Chun, SONG Gui-mei, YAN Yan, DI Peng, WANG Ying-ping. Genome Wide Identification and Expression Analysis of the bZIP Gene Family in Panax quinquefolius [J]. Biotechnology Bulletin, 2024, 40(4): 167-178. |
| [3] | LIU Huan-huan, YANG Li-chun, LI Huo-gen. Cloning and Functional Analysis of LtMYB305 in Liriodendron tulipifera [J]. Biotechnology Bulletin, 2024, 40(4): 179-188. |
| [4] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [5] | XIE Qian, JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi. Regulatory Genes Mining Related to Transcriptome Sequencing and Phenolic Metabolism Pathway of Canarium album Fruit with Different Fresh Food Quality [J]. Biotechnology Bulletin, 2024, 40(3): 215-228. |
| [6] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [7] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [8] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [9] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [10] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [11] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [12] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [13] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [14] | MEI Huan, LI Yue, LIU Ke-meng, LIU Ji-hua. Study on the Biosynthesis of l-SLR by Efficient Prokaryotic Expression of Berberine Bridge Enzyme [J]. Biotechnology Bulletin, 2023, 39(7): 277-287. |
| [15] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||