








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (2): 80-89.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0758
Previous Articles Next Articles
ZHENG Qiao1,2( ), LIN Hua1, XU Hao3, AN Wei1, XUE Chang-hua1, ZHANG Jing1(
), LIN Hua1, XU Hao3, AN Wei1, XUE Chang-hua1, ZHANG Jing1( ), HAN Guo-quan2(
), HAN Guo-quan2( )
)
Received:2023-08-09
Online:2024-02-26
Published:2024-03-13
Contact:
ZHANG Jing, HAN Guo-quan
E-mail:zq950222@163.com;zjeane@163.com;hans_980306@sicau.edu.cn
ZHENG Qiao, LIN Hua, XU Hao, AN Wei, XUE Chang-hua, ZHANG Jing, HAN Guo-quan. Establishment and Application of Multiplex Droplet Digital PCR for SARS-CoV-2 Omicron Variant[J]. Biotechnology Bulletin, 2024, 40(2): 80-89.
| 引物与探针Primer and probe | 序列Sequence(5'-3') | 产物长度Product length/bp |
|---|---|---|
| ORF1ab-F | TGTGCTAATGACCCTGT | 138 |
| ORF1ab-R | GTTTAAAAACGATTGTGCATCAG | |
| ORF1ab-P | FAM-AACTCCGCGAACCCATGCTT-BHQ1 | |
| N-F | GTCAAGCCTCTTCTCGTT | 101 |
| N-R | ATTGCCAGCCATTCTAGCAG | |
| N-P | HEX-CCTCATCACGTAGTCGCAACAGT-BHQ1 | |
| E-F | TTTCTTGCTTTCGTGGT | 132 |
| E-R | TTTAACACGAGAGTAAACGTA | |
| E-P | ROX-CTTCGATTGTGTGCGTACTGCT-BHQ2 | |
| S-F | GTTGCTGATTATTCTGTCCTA | 103 |
| S-R | AGACATTAGTAAAGCAGAGAT | |
| S-P | CY5-ACACTTAAAAGCGAAAAATGGT-MGB |
Table 1 Primer and probe sequences
| 引物与探针Primer and probe | 序列Sequence(5'-3') | 产物长度Product length/bp |
|---|---|---|
| ORF1ab-F | TGTGCTAATGACCCTGT | 138 |
| ORF1ab-R | GTTTAAAAACGATTGTGCATCAG | |
| ORF1ab-P | FAM-AACTCCGCGAACCCATGCTT-BHQ1 | |
| N-F | GTCAAGCCTCTTCTCGTT | 101 |
| N-R | ATTGCCAGCCATTCTAGCAG | |
| N-P | HEX-CCTCATCACGTAGTCGCAACAGT-BHQ1 | |
| E-F | TTTCTTGCTTTCGTGGT | 132 |
| E-R | TTTAACACGAGAGTAAACGTA | |
| E-P | ROX-CTTCGATTGTGTGCGTACTGCT-BHQ2 | |
| S-F | GTTGCTGATTATTCTGTCCTA | 103 |
| S-R | AGACATTAGTAAAGCAGAGAT | |
| S-P | CY5-ACACTTAAAAGCGAAAAATGGT-MGB |

Fig. 1 Electrophoresis map of SARS-CoV-2 ORF1ab, N, E, and S gene amplification products M: DL2000; 1: ORF1ab gene amplified product; 2: N gene amplified products; 3: E gene amplified products; 4: S gene amplified product

Fig. 2 One-dimensional map of SARS-CoV-2 ORF1ab gene, N gene, E gene and S gene by single ddPCR A: ORF1ab positive plasmid standard, blue droplets; B: N gene positive plasmid standard, red droplet; C: E gene positive plasmid standard, green droplet; D: S gene positive plasmid standard, yellow droplet; blank control, grey droplets

Fig. 3 One-dimensional map of specificity detected by quadruple ddPCR 1F: Beta strain; 1G: Gamma strain; 1H: Delta strain; 2A: SARS-CoV-2 positive plasmid standard; 2B: porcine epidemic diarrhea virus; 2C: transmissible gastroenteritis of swine virus; 2D: influenza A virus; 2E: norovirus; 2F: adenovirus; 2G: rotavirus; 2H: blank control
| 基因名称 Gene name | 稀释度 Dilution | ddPCR 检测结果Results of ddPCR/(Cp·μL-1) | 平均值AVG | 标准差SD | 相对标准偏差RSD/% | ||
|---|---|---|---|---|---|---|---|
| Repeat 1 | Repeat 2 | Repeat 3 | |||||
| ORF1ab | 10-6 | 11230.63 | 11200.05 | 11315.73 | 11248.80 | 48.94 | 0.44 |
| 10-7 | 1163.21 | 1149.84 | 1172.26 | 1161.77 | 9.21 | 0.79 | |
| 10-8 | 96.43 | 88.66 | 89.98 | 91.69 | 3.39 | 3.70 | |
| 10-9 | 8.83 | 6.57 | 7.33 | 7.58 | 0.94 | 12.39 | |
| 10-10 | 0.74 | 0.44 | 0.58 | 0.59 | 0.12 | 20.89 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |
| N | 10-6 | 6133.75 | 6226.00 | 6081.97 | 6147.21 | 59.57 | 0.97 |
| 10-7 | 617.10 | 621.28 | 598.72 | 612.37 | 9.80 | 1.60 | |
| 10-8 | 51.33 | 57.58 | 52.90 | 53.94 | 2.62 | 4.92 | |
| 10-9 | 5.33 | 5.69 | 4.40 | 5.14 | 0.54 | 10.57 | |
| 10-10 | 0.82 | 0.78 | 0.44 | 0.68 | 0.17 | 25.07 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |
| E | 10-6 | 4350.43 | 4378.88 | 4334.00 | 4387.77 | 47.95 | 1.09 |
| 10-7 | 461.63 | 443.83 | 441.59 | 449.02 | 8.97 | 2.00 | |
| 10-8 | 45.01 | 50.60 | 43.13 | 46.25 | 3.17 | 6.86 | |
| 10-9 | 4.38 | 4.87 | 4.03 | 4.43 | 0.34 | 7.78 | |
| 10-10 | 1.32 | 1.69 | 1.32 | 1.44 | 0.17 | 12.08 | |
| 10-11 | 0 | 0.44 | 0 | 0.15 | 0.21 | 141.42 | |
| S | 10-6 | 5891.09 | 5839.83 | 5942.93 | 5891.28 | 42.09 | 0.71 |
| 10-7 | 591.21 | 558.87 | 593.71 | 581.26 | 15.87 | 2.73 | |
| 10-8 | 56.59 | 55.57 | 60.52 | 57.56 | 2.13 | 3.71 | |
| 10-9 | 5.06 | 5.87 | 5.92 | 5.62 | 0.39 | 7.02 | |
| 10-10 | 0.88 | 0.88 | 1.32 | 1.03 | 0.21 | 20.20 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |
Table 2 Sensitivity and stability detection results by quadruple ddPCR
| 基因名称 Gene name | 稀释度 Dilution | ddPCR 检测结果Results of ddPCR/(Cp·μL-1) | 平均值AVG | 标准差SD | 相对标准偏差RSD/% | ||
|---|---|---|---|---|---|---|---|
| Repeat 1 | Repeat 2 | Repeat 3 | |||||
| ORF1ab | 10-6 | 11230.63 | 11200.05 | 11315.73 | 11248.80 | 48.94 | 0.44 |
| 10-7 | 1163.21 | 1149.84 | 1172.26 | 1161.77 | 9.21 | 0.79 | |
| 10-8 | 96.43 | 88.66 | 89.98 | 91.69 | 3.39 | 3.70 | |
| 10-9 | 8.83 | 6.57 | 7.33 | 7.58 | 0.94 | 12.39 | |
| 10-10 | 0.74 | 0.44 | 0.58 | 0.59 | 0.12 | 20.89 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |
| N | 10-6 | 6133.75 | 6226.00 | 6081.97 | 6147.21 | 59.57 | 0.97 |
| 10-7 | 617.10 | 621.28 | 598.72 | 612.37 | 9.80 | 1.60 | |
| 10-8 | 51.33 | 57.58 | 52.90 | 53.94 | 2.62 | 4.92 | |
| 10-9 | 5.33 | 5.69 | 4.40 | 5.14 | 0.54 | 10.57 | |
| 10-10 | 0.82 | 0.78 | 0.44 | 0.68 | 0.17 | 25.07 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |
| E | 10-6 | 4350.43 | 4378.88 | 4334.00 | 4387.77 | 47.95 | 1.09 |
| 10-7 | 461.63 | 443.83 | 441.59 | 449.02 | 8.97 | 2.00 | |
| 10-8 | 45.01 | 50.60 | 43.13 | 46.25 | 3.17 | 6.86 | |
| 10-9 | 4.38 | 4.87 | 4.03 | 4.43 | 0.34 | 7.78 | |
| 10-10 | 1.32 | 1.69 | 1.32 | 1.44 | 0.17 | 12.08 | |
| 10-11 | 0 | 0.44 | 0 | 0.15 | 0.21 | 141.42 | |
| S | 10-6 | 5891.09 | 5839.83 | 5942.93 | 5891.28 | 42.09 | 0.71 |
| 10-7 | 591.21 | 558.87 | 593.71 | 581.26 | 15.87 | 2.73 | |
| 10-8 | 56.59 | 55.57 | 60.52 | 57.56 | 2.13 | 3.71 | |
| 10-9 | 5.06 | 5.87 | 5.92 | 5.62 | 0.39 | 7.02 | |
| 10-10 | 0.88 | 0.88 | 1.32 | 1.03 | 0.21 | 20.20 | |
| 10-11 | 0 | 0 | 0 | 0 | 0 | / | |

Fig. 4 One-dimensional map of sensitivity detected by quadruple ddPCR A: FAM channel, one-dimensional map of ORF1ab gene detection; B: HEX channel, one-dimensional map of N-gene detection; C: ROX channel, one-dimensional map of E gene detection; D: CY5 channel, one-dimensional map of S gene detection
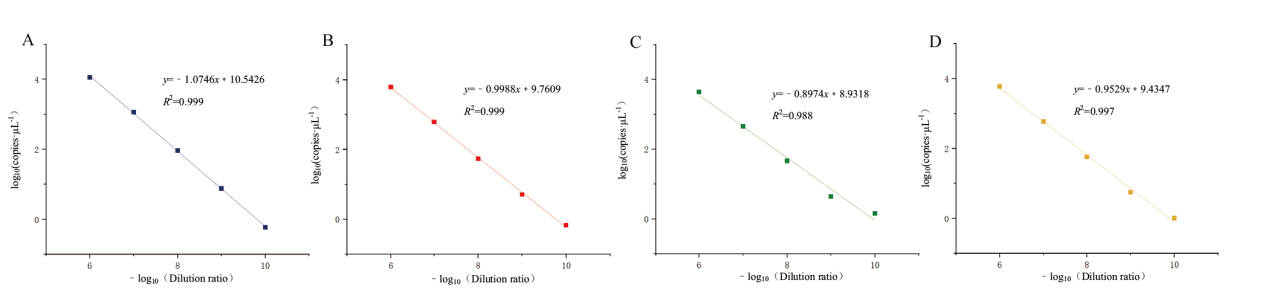
Fig. 5 Fitting curve of linear relationship for quadruple ddPCR sensitivity detection A: The fitting curve of the linear relationship of the sensitivity detection of ORF1ab gene. B: The fitting curve of the linear relationship of the sensitivity detection of N gene.C: The fitting curve of the linear relationship of the sensitivity detection of E gene. D: The fitting curve of the linear relationship of the sensitivity detection of S gene
| 样品名称No. of samples | 总微滴数Total number of droplets | ORF1ab/(Cp·μL-1) | N/(Cp·μL-1) | E/(Cp·μL-1) | S/(Cp·μL-1) |
|---|---|---|---|---|---|
| 样本1 | 20891 | 47.96 | 7480.95 | 335.57 | 99.29 |
| 样本2 | 21153 | 9.97 | 887.71 | 17.38 | 4.33 |
| 样本3 | 21372 | 4.70 | 219.61 | 9.71 | 6.45 |
| 样本4 | 20833 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本5 | 20536 | 28.03 | 6458.03 | 276.39 | 138.53 |
| 样本6 | 21393 | 14.59 | 912.05 | 21.85 | 11.59 |
| 样本7 | 21106 | 9.09 | 234.45 | 13.01 | 2.64 |
| 样本8 | 20712 | 248.53 | 7949.99 | 399.37 | 305.53 |
| 样本9 | 20793 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本10 | 21397 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本11 | 21368 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本12 | 21090 | 8.73 | 143.22 | 24.77 | 6.09 |
| 样本13 | 20675 | 5.57 | 112.42 | 19.40 | 2.20 |
| 样本14 | 20874 | 3.96 | 24.64 | 2.24 | 1.49 |
| 样本15 | 21704 | 93.39 | 6887.53 | 378.12 | 84.85 |
| 样本16 | 20380 | 38.92 | 780.27 | 357.50 | 21.19 |
| 样本17 | 21228 | 26.01 | 226.01 | 285.21 | 9.09 |
| 样本18 | 21104 | 29.11 | 269.19 | 14.39 | 24.35 |
| 样本19 | 20930 | 4.08 | 62.85 | 5.20 | 1.76 |
| 样本20 | 20678 | 118.65 | 10636.56 | 342.67 | 113.30 |
Table 3 Copy number detection results for 20 samples by quadruple ddPCR
| 样品名称No. of samples | 总微滴数Total number of droplets | ORF1ab/(Cp·μL-1) | N/(Cp·μL-1) | E/(Cp·μL-1) | S/(Cp·μL-1) |
|---|---|---|---|---|---|
| 样本1 | 20891 | 47.96 | 7480.95 | 335.57 | 99.29 |
| 样本2 | 21153 | 9.97 | 887.71 | 17.38 | 4.33 |
| 样本3 | 21372 | 4.70 | 219.61 | 9.71 | 6.45 |
| 样本4 | 20833 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本5 | 20536 | 28.03 | 6458.03 | 276.39 | 138.53 |
| 样本6 | 21393 | 14.59 | 912.05 | 21.85 | 11.59 |
| 样本7 | 21106 | 9.09 | 234.45 | 13.01 | 2.64 |
| 样本8 | 20712 | 248.53 | 7949.99 | 399.37 | 305.53 |
| 样本9 | 20793 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本10 | 21397 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本11 | 21368 | 0.00 | 0.00 | 0.00 | 0.00 |
| 样本12 | 21090 | 8.73 | 143.22 | 24.77 | 6.09 |
| 样本13 | 20675 | 5.57 | 112.42 | 19.40 | 2.20 |
| 样本14 | 20874 | 3.96 | 24.64 | 2.24 | 1.49 |
| 样本15 | 21704 | 93.39 | 6887.53 | 378.12 | 84.85 |
| 样本16 | 20380 | 38.92 | 780.27 | 357.50 | 21.19 |
| 样本17 | 21228 | 26.01 | 226.01 | 285.21 | 9.09 |
| 样本18 | 21104 | 29.11 | 269.19 | 14.39 | 24.35 |
| 样本19 | 20930 | 4.08 | 62.85 | 5.20 | 1.76 |
| 样本20 | 20678 | 118.65 | 10636.56 | 342.67 | 113.30 |
| 样品名称 No. of samples | qPCR 检测结果(ct值)qPCR results | ||
|---|---|---|---|
| FAM(ORF1ab) | HEX(N) | ROX(E) | |
| 样本1 | 29.63 | 25.62 | 28.91 |
| 样本2 | 32.06 | 28.29 | 31.70 |
| 样本3 | 33.06 | 29.55 | 32.73 |
| 样本4 | NA | NA | NA |
| 样本5 | 30.63 | 26.95 | 29.49 |
| 样本6 | 31.61 | 28.17 | 30.55 |
| 样本7 | 32.75 | 29.41 | 31.78 |
| 样本8 | 27.36 | 25.29 | 25.46 |
| 样本9 | NA | NA | NA |
| 样本10 | NA | NA | NA |
| 样本11 | NA | NA | NA |
| 样本12 | 31.45 | 30.18 | 30.23 |
| 样本13 | 32.18 | 30.89 | 30.75 |
| 样本14 | 34.35 | 33.91 | 35.34 |
| 样本15 | 28.47 | 26.79 | 26.82 |
| 样本16 | 29.86 | 28.24 | 28.2 |
| 样本17 | 30.83 | 29.40 | 29.42 |
| 样本18 | 30.83 | 29.23 | 31.17 |
| 样本19 | 33.00 | 31.68 | 33.53 |
| 样本20 | 28.29 | 24.97 | 28.78 |
Table 4 Results of 20 samples via fluorescence quantitative PCR
| 样品名称 No. of samples | qPCR 检测结果(ct值)qPCR results | ||
|---|---|---|---|
| FAM(ORF1ab) | HEX(N) | ROX(E) | |
| 样本1 | 29.63 | 25.62 | 28.91 |
| 样本2 | 32.06 | 28.29 | 31.70 |
| 样本3 | 33.06 | 29.55 | 32.73 |
| 样本4 | NA | NA | NA |
| 样本5 | 30.63 | 26.95 | 29.49 |
| 样本6 | 31.61 | 28.17 | 30.55 |
| 样本7 | 32.75 | 29.41 | 31.78 |
| 样本8 | 27.36 | 25.29 | 25.46 |
| 样本9 | NA | NA | NA |
| 样本10 | NA | NA | NA |
| 样本11 | NA | NA | NA |
| 样本12 | 31.45 | 30.18 | 30.23 |
| 样本13 | 32.18 | 30.89 | 30.75 |
| 样本14 | 34.35 | 33.91 | 35.34 |
| 样本15 | 28.47 | 26.79 | 26.82 |
| 样本16 | 29.86 | 28.24 | 28.2 |
| 样本17 | 30.83 | 29.40 | 29.42 |
| 样本18 | 30.83 | 29.23 | 31.17 |
| 样本19 | 33.00 | 31.68 | 33.53 |
| 样本20 | 28.29 | 24.97 | 28.78 |
| [1] |
Shereen MA, Khan S, Kazmi A, et al. COVID-19 infection: emergence, transmission, and characteristics of human coronaviruses[J]. J Adv Res, 2020, 24: 91-98.
doi: 10.1016/j.jare.2020.03.005 URL |
| [2] |
李家俊, 郑潇, 盛杰, 等. 新型冠状病毒及其临床检测方法研究进展[J]. 生物技术通报, 2021, 37(4): 282-292.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0954 |
| Li JJ, Zheng X, Sheng J, et al. Novel coronavirus and research progress of related clinical detection methods[J]. Biotechnol Bull, 2021, 37(4): 282-292. | |
| [3] |
Andryukov BG, Besednova NN, Kuznetsova TA, et al. Laboratory-based resources for COVID-19 diagnostics: traditional tools and novel technologies. A perspective of personalized medicine[J]. J Pers Med, 2021, 11(1): 42.
doi: 10.3390/jpm11010042 URL |
| [4] |
Chan JFW, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan[J]. Emerg Microbes Infect, 2020, 9(1): 221-236.
doi: 10.1080/22221751.2020.1719902 URL |
| [5] |
Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin[J]. Nature, 2020, 579(7798): 270-273.
doi: 10.1038/s41586-020-2012-7 |
| [6] | Bal A, Destras G, Gaymard A, et al. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020[J]. Euro Surveill, 2021, 26(3): 2100008. |
| [7] |
Chen JH, Wang R, Wang ML, et al. Mutations strengthened SARS-CoV-2 infectivity[J]. J Mol Biol, 2020, 432(19): 5212-5226.
doi: S0022-2836(20)30456-3 pmid: 32710986 |
| [8] |
Dejnirattisai W, Shaw RH, Supasa P, et al. Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum[J]. Lancet, 2022, 399(10321): 234-236.
doi: 10.1016/S0140-6736(21)02844-0 URL |
| [9] | Feder KA, Pearlowitz M, Goode A, et al. Linked clusters of SARS-CoV-2 variant B.1.351 - Maryland, january-february 2021[J]. Morb Mortal Wkly Rep, 2021, 70(17): 627-631. |
| [10] | Galloway SE, Paul P, MacCannell DR, et al. Emergence of SARS-CoV-2 B.1.1.7 lineage - United States, December 29, 2020-january 12, 2021[J]. Morb Mortal Wkly Rep, 2021, 70(3): 95-99. |
| [11] | 孙泽宇, 柴佳彤, 许建成. 新冠病毒变异株“奥密克戎”的研究进展[J]. 病毒学报, 2023, 39(2): 517-527. |
| Sun ZY, Chai JT, Xu JC. Research progress on the Omicron variant of SARS-CoV-2[J]. Chin J Virol, 2023, 39(2): 517-527. | |
| [12] |
Coutard B, Valle C, de Lamballerie X, et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade[J]. Antiviral Res, 2020, 176: 104742.
doi: 10.1016/j.antiviral.2020.104742 URL |
| [13] |
Benton DJ, Wrobel AG, Xu PQ, et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion[J]. Nature, 2020, 588(7837): 327-330.
doi: 10.1038/s41586-020-2772-0 |
| [14] |
Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR[J]. Nat Methods, 2013, 10(10): 1003-1005.
doi: 10.1038/nmeth.2633 pmid: 23995387 |
| [15] |
Li HY, Bai RL, Zhao ZY, et al. Application of droplet digital PCR to detect the pathogens of infectious diseases[J]. Biosci Rep, 2018, 38(6): BSR20181170.
doi: 10.1042/BSR20181170 URL |
| [16] | 王志彬, 罗庆, 李宝林, 等. 数字PCR技术检测感染者尿液中SARS-CoV-2的方法研究[J]. 西南医科大学学报, 2022, 45(4): 301-307. |
| Wang ZB, Luo Q, Li BL, et al. Study on the detection of SARS-CoV-2 in the urine of infected persons by droplet digital PCR[J]. J Southwest Med Univ, 2022, 45(4): 301-307. | |
| [17] | 杨朔鹏. 微滴式数字PCR用于生物与环境样本中SARS-CoV-2的高灵敏检测[D]. 石家庄: 河北医科大学, 2022. |
| Yang SP. Droplet digital PCR was used for the highly sensitive detection of SARS-CoV-2 in biological and environmental samples[D]. Shijiazhuang: Hebei Medical University, 2022. | |
| [18] |
Zhang Z, Wang N, Liu XF, et al. A novel, reverse transcription, droplet digital PCR assay for the combined, sensitive detection of severe acute respiratory syndrome coronavirus 2 with swine acute diarrhea syndrome coronavirus[J]. J AOAC Int, 2022, 105(5): 1437-1446.
doi: 10.1093/jaoacint/qsac039 pmid: 35377440 |
| [19] |
Collie S, Champion J, Moultrie H, et al. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa[J]. N Engl J Med, 2022, 386(5): 494-496.
doi: 10.1056/NEJMc2119270 URL |
| [20] |
Ren WL, Zhang Y, Rao JH, et al. Evolution of immune evasion and host range expansion by the SARS-CoV-2 B.1.1.529(Omicron)variant[J]. mBio, 2023, 14(2): e0041623.
doi: 10.1128/mbio.00416-23 URL |
| [21] |
Qu PK, Evans JP, Faraone JN, et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2[J]. Cell Host Microbe, 2023, 31(1): 9-17.e3.
doi: 10.1016/j.chom.2022.11.012 URL |
| [22] |
Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus(2019-nCoV)causing an outbreak of pneumonia[J]. Clin Chem, 2020, 66(4): 549-555.
doi: 10.1093/clinchem/hvaa029 URL |
| [23] | Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus(2019-nCoV)by real-time RT-PCR[J]. Euro Surveill, 2020, 25(3): 2000045. |
| [24] | 张稳健, 吕欣, 黄驰, 等. 胶体金免疫层析法检测新型冠状病毒IgM/IgG抗体的临床评价与应用[J]. 病毒学报, 2020, 36(3): 348-354. |
| Zhang WJ, Lyu X, Huang C, et al. Clinical evaluation and application of detection of IgM and IgG antibodies against SARS-CoV-2 using a colloidal gold immunochromatography assay[J]. Chin J Virol, 2020, 36(3): 348-354. | |
| [25] |
Cremonesi P, Cortimiglia C, Picozzi C, et al. Development of a droplet digital polymerase chain reaction for rapid and simultaneous identification of common foodborne pathogens in soft cheese[J]. Front Microbiol, 2016, 7: 1725.
pmid: 27840628 |
| [1] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [2] | YU Hui, WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun. Isolation and Functional Verification of Genes Responding to Iron and Cadmium Stresses in Lycium barbarum [J]. Biotechnology Bulletin, 2023, 39(7): 195-205. |
| [3] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [4] | SONG Hai-na, WU Xin-tong, YANG Lu-yu, GENG Xi-ning, ZHANG Hua-min, SONG Xiao-long. Selection and Validation of Reference Genes for RT-qPCR in Allium tuberosum Infected by Botrytis squamosa [J]. Biotechnology Bulletin, 2023, 39(3): 101-115. |
| [5] | LI Yi-ya, WU Yi-fan, DING Neng-shui, FAN Xiao-ping, CHEN Fan. Establishment of a Luciferase-assisted Quantitative Method for Measuring Ultrasonic Disruption of Escherichia coli Cells [J]. Biotechnology Bulletin, 2023, 39(12): 90-98. |
| [6] | LI Hui-jie, DONG Lian-hua, CHEN Gui-fang, LIU Si-yuan, YANG Jia-yi, YANG Jing-ya. Establishment of Droplet Digital PCR Assay for Quantitative Detection of Pseudomonas cocovenenans in Foods [J]. Biotechnology Bulletin, 2023, 39(1): 127-136. |
| [7] | HU Xue-ying, ZHANG Yue, GUO Ya-jie, QIU Tian-lei, GAO Min, SUN Xing-bin, WANG Xu-ming. Comparison in Antibiotic Resistance Genes Carried by Bacteriophages and Bacteria in Farmland Soil Amended with Different Fertilizers [J]. Biotechnology Bulletin, 2022, 38(9): 116-126. |
| [8] | CHENG Shen-wei, ZHANG Ke-qiang, LIANG Jun-feng, LIU Fu-yuan, GAO Xing-liang, DU Lian-zhu. Establishment of a Triple Droplet Digital PCR Quantitative Detection Method for Typical Pathogenic Bacteria in Livestock and Poultry Manure [J]. Biotechnology Bulletin, 2022, 38(9): 271-280. |
| [9] | CAO Ying-fang, ZHAO Xin, LIU Shuang, LI Rui-huan, LIU Na, XU Shi-yong, GAO Fang-rui, MA Hui, LAN Qing-kuo, TAN Jian-xin, WANG Yong. Establishment of Real-time Fluorescent Quantitative PCR Detection Method for Genetically Modified Herbicide-tolerant Soybean GE-J12 [J]. Biotechnology Bulletin, 2022, 38(7): 146-152. |
| [10] | WANG Jia-li, HE Si-qi, KANG Zi-xi, WANG Jian-xun. Antibody Phage Display Technology and Its Application in the Discovery of Anti-SARS-CoV-2 Antibodies [J]. Biotechnology Bulletin, 2022, 38(5): 248-256. |
| [11] | WANG Qiao-ju, HU Yu-meng, WEN Ya-ya, SONG Li, MENG Chuang, PAN Zhi-ming, JIAO Xin-an. Expression and Activity Identification of SARS-CoV-2 S1 Protein [J]. Biotechnology Bulletin, 2022, 38(3): 157-163. |
| [12] | LIU Xiao-mei, WANG Dong-xin, ZHANG Chun, WEI Shuang-shi. Inhibition of AAV-mediated RNAi to SARS-CoV-2 S Gene Expression [J]. Biotechnology Bulletin, 2022, 38(3): 188-193. |
| [13] | SU Yuan, ZHU Long-jiao, CAO Ji-juan, LIU Jian-long, XU Wen-tao. Development of Fluorescence Quantitative Lyophilized Detection Kit Based on Escherichia coli O157∶H7 [J]. Biotechnology Bulletin, 2022, 38(3): 264-275. |
| [14] | LAN Xin-yue, LIU Ning-ning, ZHU Long-jiao, CHEN Xu, CHU Hua-shuo, LI Xiang-yang, DUAN Nuo, XU Wen-tao. Tetracycline Bivalent Aptamer Non-enzyme Label-free Sensor [J]. Biotechnology Bulletin, 2022, 38(3): 276-284. |
| [15] | CHEN Duo, LIU Yong-zhe. Prokaryotic Expression,Purification and Crystallization of N-terminal Domain of Nucleocapsid Protein in SARS-CoV-2 [J]. Biotechnology Bulletin, 2022, 38(12): 149-155. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||