








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (5): 310-320.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1114
Previous Articles Next Articles
WANG Xin-xin1,2( ), GUAN Yu-zhu1,2, LI Xiao-wei1,2, HONG Wei1,3, WU Dao-yan1,2, KANG Ying-qian1,2, LIU Yong-chang1,2, CHEN Zheng-hong1,2(
), GUAN Yu-zhu1,2, LI Xiao-wei1,2, HONG Wei1,3, WU Dao-yan1,2, KANG Ying-qian1,2, LIU Yong-chang1,2, CHEN Zheng-hong1,2( ), CUI Gu-zhen1,2(
), CUI Gu-zhen1,2( )
)
Received:2023-11-27
Online:2024-05-26
Published:2024-03-28
Contact:
CHEN Zheng-hong, CUI Gu-zhen
E-mail:wxx_999w@163.com;chenzhenghong@gmc.edu.cn;cuiguzhen@gmc.edu.cn
WANG Xin-xin, GUAN Yu-zhu, LI Xiao-wei, HONG Wei, WU Dao-yan, KANG Ying-qian, LIU Yong-chang, CHEN Zheng-hong, CUI Gu-zhen. Function of katA in Helicobacter pylori and Its Role in the Tolerance to Oxidative Damage[J]. Biotechnology Bulletin, 2024, 40(5): 310-320.
| 菌株及质粒Strain and plasmid | 相关特征Relevant characteristics | 来源Source |
|---|---|---|
| Strains E.coli | ||
| DH5α | Clone strain, F-lacZ ΔM15Δ(lacZYA-aragF)relA1 | This lab |
| DH5α::pSD | Derived fom DH5α, carrying plasmid pSD, kanR | This lab |
| DH5α:: pSD-KO-katA | Derived fom DH5α, carrying plasmid pSD-KO-katA, kanR | This work |
| DH5α:: pCD | Derived fom DH5α, carrying plasmid pCD, CmR | This work |
| DH5α:: pCD-EX-katA | Derived fom DH5α, carrying plasmid pCD-EX-katA, CmR | This work |
| DH5α:: pET-28a-katA | Derived fom DH5α, carrying plasmid pET-28a-katA, kanR | This work |
| BL21(DE3) | Expression strain, F-lon-11Δ(ompT-nfrA)885Δ(galM-ybhJ)884 | This lab |
| BL21(DE3)::pET-28a | Derived fom BL21(DE3), carrying plasmid pET-28a, kanR | This work |
| BL21(DE3)::pET-28a-katA | Derived fom BL21(DE3), carrying plasmid pET-28a-katA, kanR | This work |
| H.pylori | ||
| HpG272 | Clinical strains | This lab |
| Hp∆katA | H.pylori strain G272, knockout gene katA | This work |
| Hp∆katA::katA | H.pylori strain HpΔkatA::katA, anaplerosis gene katA | This work |
| Plasmids | ||
| pSD | From pill570, suicide plasmid, kanR | This lab |
| PCD | From pSD, suicide plasmid, CmR | This lab |
| pSD-KO-katA | Suicide plasmid, knockout gene katA, kanR | This work |
| pCD-EX-katA | HpΔkatA::katA replenishing plasmid, CmR | This work |
| pET-28a | N-T7, N-His, C-His, E. coli protein expression vector | This lab |
| pET-28a-katA | Carrying katA gene, E. coli protein expression vector | This work |
Table 1 Strains and plasmids used in this study
| 菌株及质粒Strain and plasmid | 相关特征Relevant characteristics | 来源Source |
|---|---|---|
| Strains E.coli | ||
| DH5α | Clone strain, F-lacZ ΔM15Δ(lacZYA-aragF)relA1 | This lab |
| DH5α::pSD | Derived fom DH5α, carrying plasmid pSD, kanR | This lab |
| DH5α:: pSD-KO-katA | Derived fom DH5α, carrying plasmid pSD-KO-katA, kanR | This work |
| DH5α:: pCD | Derived fom DH5α, carrying plasmid pCD, CmR | This work |
| DH5α:: pCD-EX-katA | Derived fom DH5α, carrying plasmid pCD-EX-katA, CmR | This work |
| DH5α:: pET-28a-katA | Derived fom DH5α, carrying plasmid pET-28a-katA, kanR | This work |
| BL21(DE3) | Expression strain, F-lon-11Δ(ompT-nfrA)885Δ(galM-ybhJ)884 | This lab |
| BL21(DE3)::pET-28a | Derived fom BL21(DE3), carrying plasmid pET-28a, kanR | This work |
| BL21(DE3)::pET-28a-katA | Derived fom BL21(DE3), carrying plasmid pET-28a-katA, kanR | This work |
| H.pylori | ||
| HpG272 | Clinical strains | This lab |
| Hp∆katA | H.pylori strain G272, knockout gene katA | This work |
| Hp∆katA::katA | H.pylori strain HpΔkatA::katA, anaplerosis gene katA | This work |
| Plasmids | ||
| pSD | From pill570, suicide plasmid, kanR | This lab |
| PCD | From pSD, suicide plasmid, CmR | This lab |
| pSD-KO-katA | Suicide plasmid, knockout gene katA, kanR | This work |
| pCD-EX-katA | HpΔkatA::katA replenishing plasmid, CmR | This work |
| pET-28a | N-T7, N-His, C-His, E. coli protein expression vector | This lab |
| pET-28a-katA | Carrying katA gene, E. coli protein expression vector | This work |
| 引物Primer | 序列Sequence(5'-3') | 注释Notes |
|---|---|---|
| Kan-1 | GAATTCGAGCTCGGTACCCG | kanR 基因扩增和检测引物 |
| Kan-2 | CCCGGGTCATTATTCCCTCC | |
| Kat-1 | CTGCAGGGTATAAAAACTCACCGCCCCA | 敲除质粒及敲除菌株检测引物 |
| Kat-2 | ATCGATACCTAGTTTCAAGCCTTGCA | |
| KatA-1 | ATCGACTGCAGGGTATAAAAACTCACCGCCCCA | katA 基因敲除载体上游同源臂扩增 |
| KatA-2 | ATCGAGAATTCCGTAATTGACACTAAGCCGATTAC | |
| KatA-3 | ATCGAGGATCCCTTATTTTTTAGGAACGCTTTGTC | katA 基因敲除载体下游同源臂扩增 |
| KatA-4 | ATCGAATCGATACCTAGTTTCAAGCCTTGCA | |
| Chl-1 | TTAAAAAAATTACGCCCCGCCCTG | CmR 基因扩增和检测引物 |
| Chl-2 | AAATGGAGAAAAAAATCACTGGATATACCACCG | |
| KatA -5 | CCCCGGGGACCTGCAGGCCGCTTGCACGGGT | katA 基因回补载体上游同源臂 |
| KatA -6 | TTTTTTAAGGCAGTCTGCAGGCGACCAAAACCTCTCATGGAG | |
| KatA -7 | TTGACAGCTTATCATCGATGTGAGTCAGAATGTCTTTCACAACCC | katA 基因回补载体下游同源臂 |
| KatA -8 | CTCCTGAAAATCTCGGATCCGGGCGGTTTCACTGAGAAAACTT | |
| KatA -9 | CAGTGAAACCGCCCGGATCTTTATAAATTCTAAAGGGG | katA 基因回补载体Hp尿素酶启动子PureA 扩增 |
| KatA -10 | TTAGCTCCTGAAAATCTCGGATCCTTATTCTCCTATTCTTAAAGTG | |
| KatA -11 | AACACTTTAAGAATAGGAGAATAAGATGGTTAATAAAGATGTGAAACAAACTACTGC | 回补载体 katA 基因扩增 |
| KatA -12 | TTAGCTCCTGAAAATCTCGTTACTTTTTCTTTTTTGTGTGGTGCATGTCT | |
| Kat-3 | TTGACAGCTTATCATCGATGTGAGTCAGAATGTCTTTCACAACCC | 回补质粒及回补菌株检测引物 |
| Kat-4 | CCCCGGGGACCTGCAGGCCGCTTGCACGGGT | |
| KatA-R | GTCGACGGAGCTCGAATTCGTTACTTTTTCTTTTTTGTGTGGT | katA 基因扩增 |
| KatA-F | AGCAAATGGGTCGCGGATCATGGTTAATAAAGATGTGAAAC | |
| KatA-C1 | CAAAAAACCCCTCAAGACCCGTTTAG | KatAG272 蛋白表达质粒检测引物 |
| KatA-C2 | ACCGGCATACTCTGCGACATC |
Table 2 Primers used in this study
| 引物Primer | 序列Sequence(5'-3') | 注释Notes |
|---|---|---|
| Kan-1 | GAATTCGAGCTCGGTACCCG | kanR 基因扩增和检测引物 |
| Kan-2 | CCCGGGTCATTATTCCCTCC | |
| Kat-1 | CTGCAGGGTATAAAAACTCACCGCCCCA | 敲除质粒及敲除菌株检测引物 |
| Kat-2 | ATCGATACCTAGTTTCAAGCCTTGCA | |
| KatA-1 | ATCGACTGCAGGGTATAAAAACTCACCGCCCCA | katA 基因敲除载体上游同源臂扩增 |
| KatA-2 | ATCGAGAATTCCGTAATTGACACTAAGCCGATTAC | |
| KatA-3 | ATCGAGGATCCCTTATTTTTTAGGAACGCTTTGTC | katA 基因敲除载体下游同源臂扩增 |
| KatA-4 | ATCGAATCGATACCTAGTTTCAAGCCTTGCA | |
| Chl-1 | TTAAAAAAATTACGCCCCGCCCTG | CmR 基因扩增和检测引物 |
| Chl-2 | AAATGGAGAAAAAAATCACTGGATATACCACCG | |
| KatA -5 | CCCCGGGGACCTGCAGGCCGCTTGCACGGGT | katA 基因回补载体上游同源臂 |
| KatA -6 | TTTTTTAAGGCAGTCTGCAGGCGACCAAAACCTCTCATGGAG | |
| KatA -7 | TTGACAGCTTATCATCGATGTGAGTCAGAATGTCTTTCACAACCC | katA 基因回补载体下游同源臂 |
| KatA -8 | CTCCTGAAAATCTCGGATCCGGGCGGTTTCACTGAGAAAACTT | |
| KatA -9 | CAGTGAAACCGCCCGGATCTTTATAAATTCTAAAGGGG | katA 基因回补载体Hp尿素酶启动子PureA 扩增 |
| KatA -10 | TTAGCTCCTGAAAATCTCGGATCCTTATTCTCCTATTCTTAAAGTG | |
| KatA -11 | AACACTTTAAGAATAGGAGAATAAGATGGTTAATAAAGATGTGAAACAAACTACTGC | 回补载体 katA 基因扩增 |
| KatA -12 | TTAGCTCCTGAAAATCTCGTTACTTTTTCTTTTTTGTGTGGTGCATGTCT | |
| Kat-3 | TTGACAGCTTATCATCGATGTGAGTCAGAATGTCTTTCACAACCC | 回补质粒及回补菌株检测引物 |
| Kat-4 | CCCCGGGGACCTGCAGGCCGCTTGCACGGGT | |
| KatA-R | GTCGACGGAGCTCGAATTCGTTACTTTTTCTTTTTTGTGTGGT | katA 基因扩增 |
| KatA-F | AGCAAATGGGTCGCGGATCATGGTTAATAAAGATGTGAAAC | |
| KatA-C1 | CAAAAAACCCCTCAAGACCCGTTTAG | KatAG272 蛋白表达质粒检测引物 |
| KatA-C2 | ACCGGCATACTCTGCGACATC |
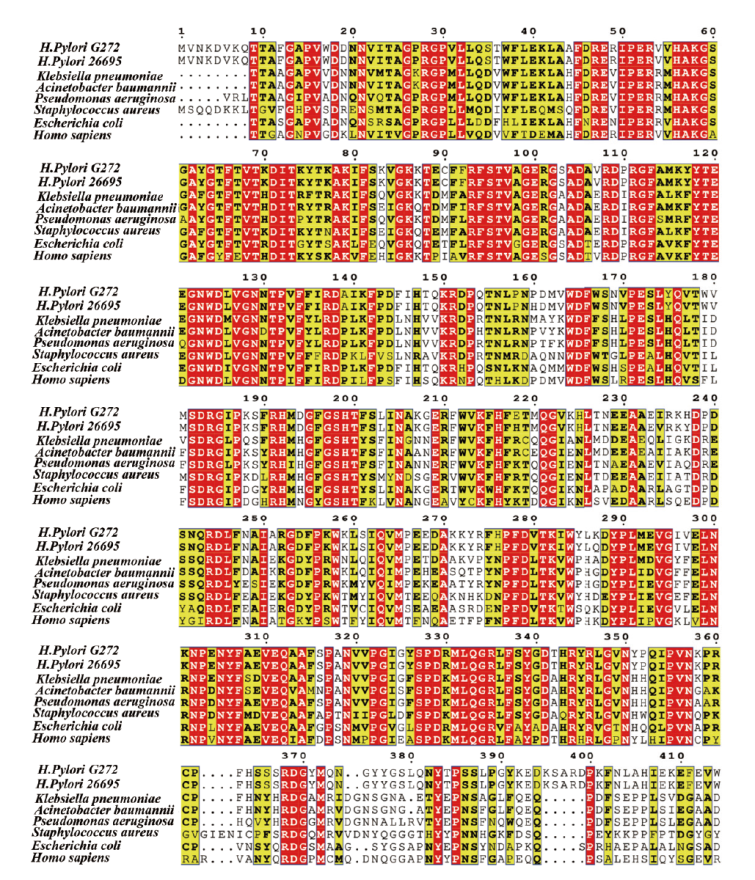
Fig. 1 Conservative analysis of amino acid sequence for katA Red: Conservative amino acids. Yellow: Relatively conserved amino acid. Colorless: Non conserved amino acids
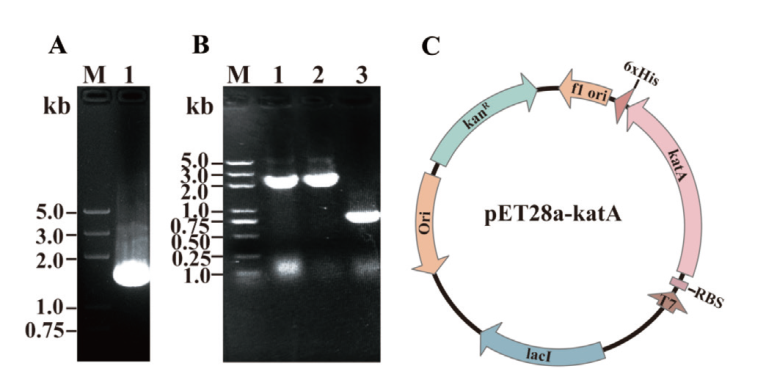
Fig. 2 PCR amplification for katA and construction of expression vector A: PCR amplification for katA gene. B: Expression plasmid PCR identification (Lane 1-2: PCR identification for positive plasmid; lane 3: blank control, primers used in this identification are KatA-C1/KatA-C2). C: pET28a-katA expression plasmid
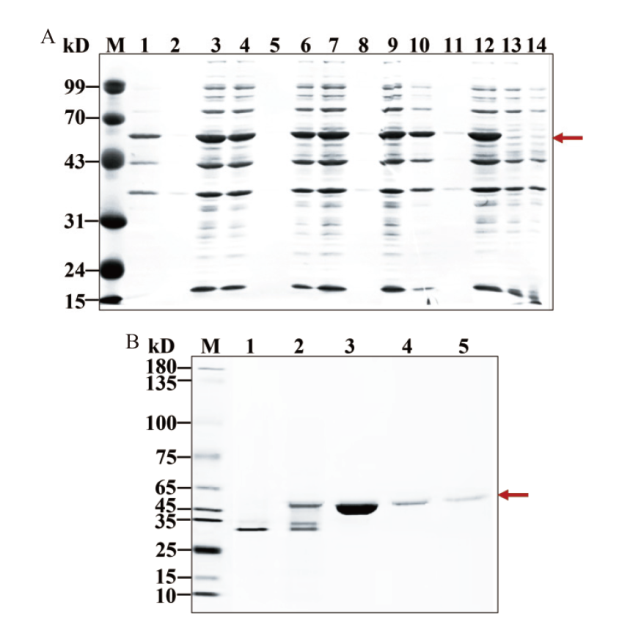
Fig. 3 Protein expression, isolation and purification for KatAG272 A: The induced expression of KatAG272 protein in Escherichia coli(Lane 1, 4, 7, 10: Bacterial precipitation before ultrasound fragmentation; lane 2, 5, 8, 11: culture supernatant before ultrasound fragmentation; lane 3, 6, 9, 12: supernatant after ultrasound fragmentation; lane 13-14: control of bacterial precipitation before wild-type ultrasound fragmentation containing pET28a vector). B: KatAG272 protein isolation and purification(Lane 1: Flow through fluid; lane 2:10 mmol/L imidazole rinse; lane 3-5: 200 mmol/L imidazole elution)
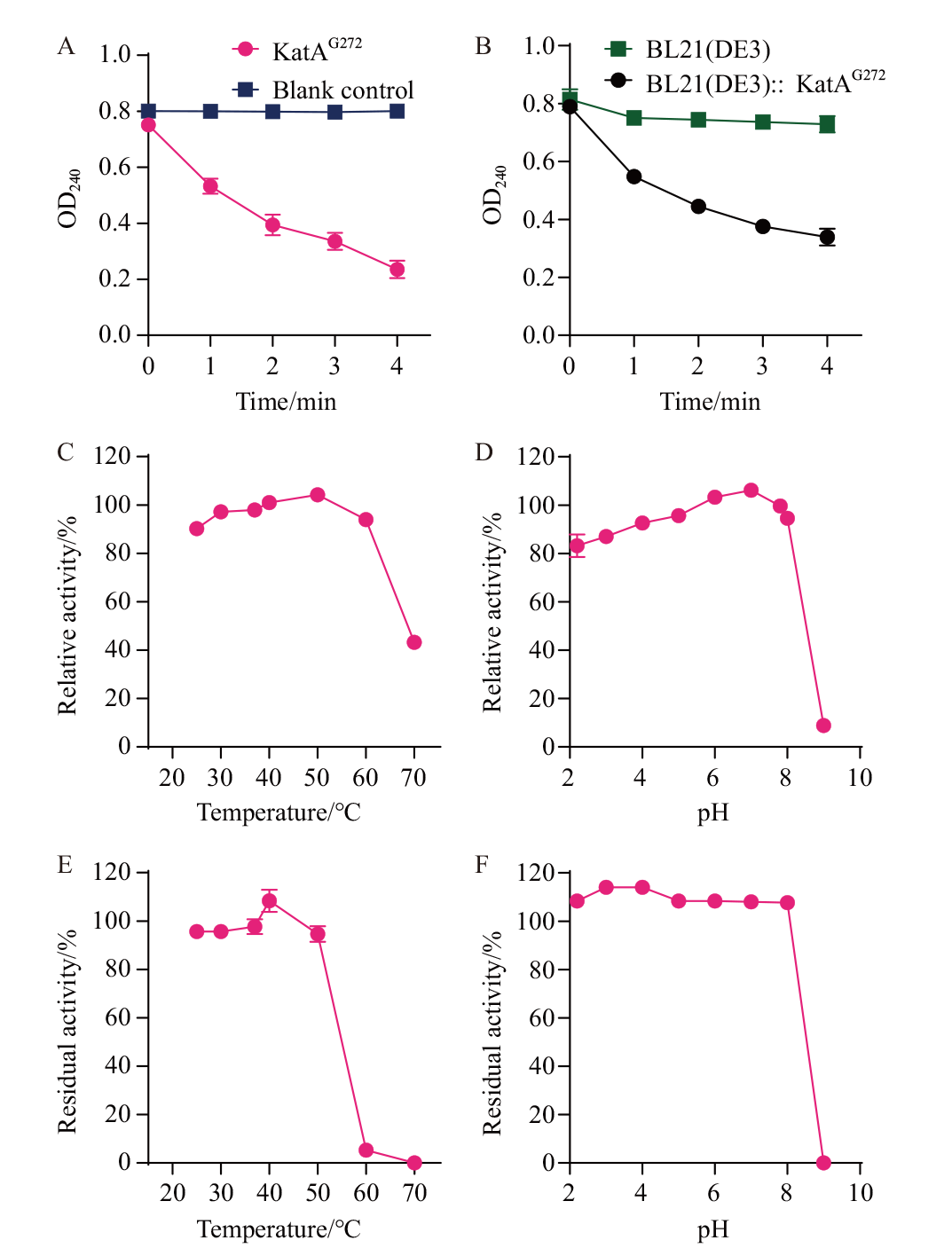
Fig. 4 Analysis and identification of catalase activity A: Detection of catalase activity of KatAG272 pure enzyme. B: Detection of KatAG272 catalase activity in recombinant E. coli; C: The effect of temperature on the activity of KatAG272. D: The effect of pH on the activity of KatAG272. E: KatAG272 protein temperature tolerance. F: KatAG272 protein pH tolerance
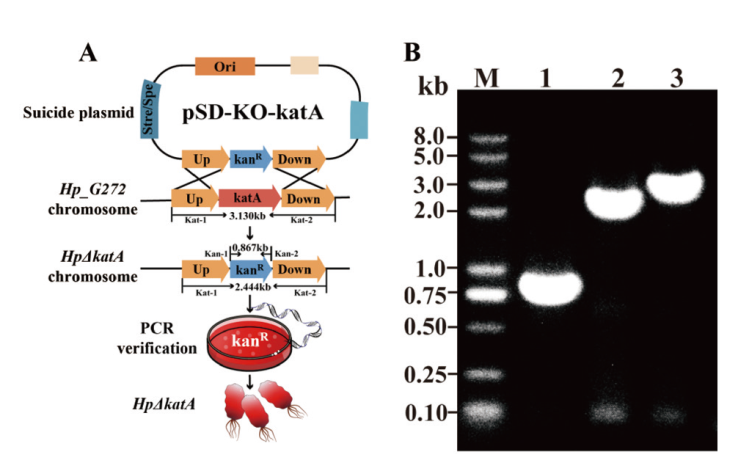
Fig. 5 Construction and identification for katA-knockout strain HpΔkatA A: Schematics of HpΔkatA construction. B: Identification of HpΔkatA(Lane 1: primer Kan-1 and Kan-2, validation of kanamycin resistance in knockout strains; lane 2: primer Kat-1 and Kat-2, validation of kanamycin resistance fragments and upstream and downstream homologous arm fragments in knockout strains; lane 3: primer Kat-1 and Kat-2, wild-type control)
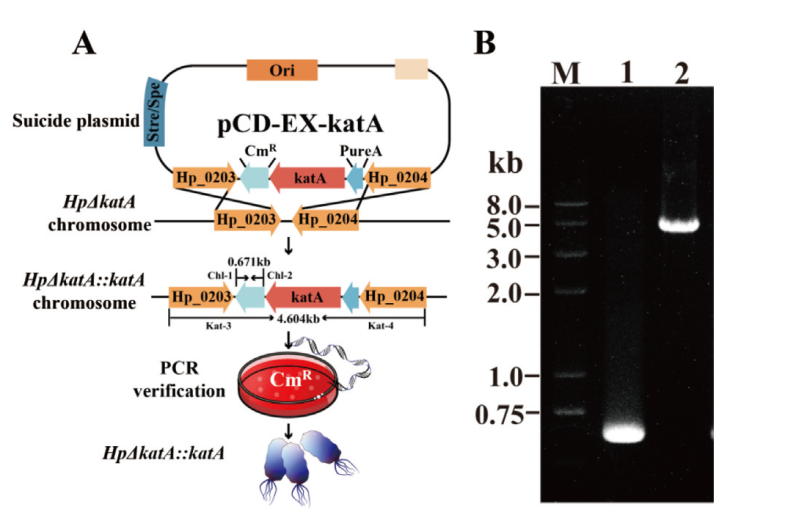
Fig. 6 Construction and identification for katA-complemented strain HpΔkatA::katA A: Schematics of HpΔkatA::katA construction; B: Identification HpΔkatA::katA(lane 1: primer Chl-1 and Chl-2, validation of chloramphenicol resistance of the complementary strain; lane 2: primer Kat-3 and Kat-4, validation of chloramphenicol resistance fragments and upstream and downstream homologous arm fragments of the complementary strain)
| 菌株Strain | 浓度Concentration/(mmol·L-1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 6 | 10 | 20 | 40 | 60 | 80 | 100 | ||
| Hp_G272 | + | + | + | + | + | + | - | - | - | |
| HpΔkatA | + | + | + | - | - | - | - | - | - | |
| HpΔkatA::katA | + | + | + | + | + | + | - | - | - | |
Table 3 Analysis of tolerance of Hp to hydrogen peroxide
| 菌株Strain | 浓度Concentration/(mmol·L-1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 6 | 10 | 20 | 40 | 60 | 80 | 100 | ||
| Hp_G272 | + | + | + | + | + | + | - | - | - | |
| HpΔkatA | + | + | + | - | - | - | - | - | - | |
| HpΔkatA::katA | + | + | + | + | + | + | - | - | - | |
| [1] |
Malfertheiner P, Camargo MC, El-Omar E, et al. Helicobacter pylori infection[J]. Nat Rev Dis Primers, 2023, 9: 19.
doi: 10.1038/s41572-023-00431-8 pmid: 37081005 |
| [2] |
Robinson K, Atherton JC. The spectrum of Helicobacter-mediated diseases[J]. Annu Rev Pathol, 2021, 16: 123-144.
doi: 10.1146/annurev-pathol-032520-024949 pmid: 33197219 |
| [3] | Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249. |
| [4] | 中国疾病预防控制中心传染病预防控制所. 中国幽门螺杆菌感染防控白皮书[M]. 北京, 2023:1-47. |
| Institute for Infectious Disease Prevention and Control. China Center for Disease Control and Prevention White Paper on Prevention and Control of Helicobacter pylori Infection in China[M]. Beijing, 2023:1-47 | |
| [5] |
Tshibangu-Kabamba E, Yamaoka Y. Helicobacter pylori infection and antibiotic resistance—from biology to clinical implications[J]. Nat Rev Gastroenterol Hepatol, 2021, 18: 613-629.
doi: 10.1038/s41575-021-00449-x pmid: 34002081 |
| [6] |
Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori[J]. Nat Rev Microbiol, 2013, 11(6): 385-399.
doi: 10.1038/nrmicro3016 pmid: 23652324 |
| [7] | Prashar A, Capurro MI, Jones NL. Under the radar: strategies used by Helicobacter pylori to evade host responses[J]. Annu Rev Physiol, 2022, 84: 485-506. |
| [8] | Sies H, Jones DP. Reactive oxygen species(ROS)as pleiotropic physiological signalling agents[J]. Nat Rev Mol Cell Biol, 2020, 21(7): 363-383. |
| [9] |
Alfonso-Prieto M, Biarnés X, Vidossich P, et al. The molecular mechanism of the catalase reaction[J]. J Am Chem Soc, 2009, 131(33): 11751-11761.
doi: 10.1021/ja9018572 pmid: 19653683 |
| [10] | Miyashita M, Joh T, Watanabe K, et al. Immune responses in mice to intranasal and intracutaneous administration of a DNA vaccine encoding Helicobacter pylori-catalase[J]. Vaccine, 2002, 20(17/18): 2336-2342. |
| [11] |
Radcliff FJ, Hazell SL, Kolesnikow T, et al. Catalase, a novel antigen for Helicobacter pylori vaccination[J]. Infect Immun, 1997, 65(11): 4668-4674.
doi: 10.1128/iai.65.11.4668-4674.1997 pmid: 9353048 |
| [12] | Harris AG, Wilson JE, Danon SJ, et al. Catalase(KatA)and KatA-associated protein(KapA)are essential to persistent colonization in the Helicobacter pylori SS1 mouse model[J]. Microbiology, 2003, 149(Pt 3): 665-672. |
| [13] | Harris AG, Hinds FE, Beckhouse AG, et al. Resistance to hydrogen peroxide in Helicobacter pylori: role of catalase(KatA)and Fur, and functional analysis of a novel gene product designated ‘KatA-associated protein', KapA(HP0874)[J]. Microbiology, 2002, 148(Pt 12): 3813-3825. |
| [14] | Cardenas VM, Dominguez DC, Puentes FA, et al. Evaluation of a novel stool native catalase antigen test for Helicobacter pylori infection in asymptomatic North American children[J]. J Pediatr Gastroenterol Nutr, 2008, 46(4): 399-402. |
| [15] | Okuda M, Osaki T, Kikuchi S, et al. Evaluation of a stool antigen test using a MAb for native catalase for diagnosis of Helicobacter pylori infection in children and adults[J]. J Med Microbiol, 2014, 63(Pt 12): 1621-1625. |
| [16] |
Shimoyama T, Sawaya M, Ishiguro A, et al. Applicability of a rapid stool antigen test, using monoclonal antibody to catalase, for the management of Helicobacter pylori infection[J]. J Gastroenterol, 2011, 46(4): 487-491.
doi: 10.1007/s00535-011-0371-4 pmid: 21264478 |
| [17] | Sato M, Shimoyama T, Takahashi R, et al. Characterization and usefulness of stool antigen tests using a monoclonal antibody to Helicobacter pylori catalase[J]. J Gastroenterol Hepatol, 2012, 27(Suppl 3): 23-28. |
| [18] | Zhang B, Li HL, Fan Q, et al. Serum Helicobacter pylori KatA and AhpC antibodies as novel biomarkers for gastric cancer[J]. World J Gastroenterol, 2016, 22(21): 5060-5067. |
| [19] | Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server[J]. Nucleic Acids Res, 2014, 42(Web Server issue): W320-W324. |
| [20] |
Yoshida N, Granger DN, Evans DJ Jr, et al. Mechanisms involved in Helicobacter pylori-induced inflammation[J]. Gastroenterology, 1993, 105(5): 1431-1440.
pmid: 7901109 |
| [21] | Basu M, Czinn SJ, Blanchard TG. Absence of catalase reduces long-term survival of Helicobacter pylori in macrophage phagosomes[J]. Helicobacter, 2004, 9(3): 211-216. |
| [22] |
Ramarao N, Gray-Owen SD, Meyer TF. Helicobacter pylori induces but survives the extracellular release of oxygen radicals from professional phagocytes using its catalase activity[J]. Mol Microbiol, 2000, 38(1): 103-113.
pmid: 11029693 |
| [23] | Richter C, Mukherjee O, Ermert D, et al. Moonlighting of Helicobacter pylori catalase protects against complement-mediated killing by utilising the host molecule vitronectin[J]. Sci Rep, 2016, 6: 24391. |
| [24] | Usui Y, Taniyama Y, Endo M, et al. Helicobacter pylori, homologous-recombination genes, and gastric cancer[J]. N Engl J Med, 2023, 388(13): 1181-1190. |
| [25] | Shah SC, Tepler A, Chung CP, et al. Host genetic determinants associated with Helicobacter pylori eradication treatment failure: a systematic review and meta-analysis[J]. Gastroenterology, 2021, 161(5): 1443-1459. |
| [26] | Muñoz-Ramirez ZY, Pascoe B, Mendez-Tenorio A, et al. A 500-year tale of co-evolution, adaptation, and virulence: Helicobacter pylori in the Americas[J]. ISME J, 2021, 15: 78-92. |
| [27] |
Palamides P, Jolaiya T, Idowu A, et al. Helicobacter pylori patient isolates from South Africa and Nigeria differ in virulence factor pathogenicity profile and associated gastric disease outcome[J]. Sci Rep, 2020, 10(1): 11409.
doi: 10.1038/s41598-020-66128-0 pmid: 32651394 |
| [28] | Mi MH, Wu FC, Zhu J, et al. Heterogeneity of Helicobacter pylori strains isolated from patients with gastric disorders in Guiyang, China[J]. Infect Drug Resist, 2021, 14: 535-545. |
| [29] | Hosoda K, Wanibuchi K, Amgalanbaatar A, et al. A novel role of catalase in cholesterol uptake of Helicobacter pylori[J]. Steroids, 2023, 191: 109158. |
| [30] | Zhang Y, Li XY, Shan BE, et al. Perspectives from recent advances of Helicobacter pylori vaccines research[J]. Helicobacter, 2022, 27(6): e12926. |
| [1] | ZHOU Hong-dan, LUO Xiao-ping, TU Mi-xue, LI Zhong-guang. Phytomelatonin: An Emerging Signal Molecule Responding to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(3): 41-51. |
| [2] | GAO Deng-ke, MA Bai-rong, GUO Yi-ying, LIU Wei, LIU Tian, JIN Ya-ping, JIANG Zhou, CHEN Hua-tao. Establishment of Quaking Knockout Mouse Embryonic Fibroblast Cell Line Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2024, 40(2): 65-72. |
| [3] | WANG Yu-qing, MA Zi-qi, HOU Jia-xin, ZONG Yu-qi, HAO Han-rui, LIU Guo-yuan, WEI Hui, LIAN Bo-lin, CHEN Yan-hong, ZHANG Jian. Research Progress in the Composition Analysis and Ecological Function of Plant Root Exudates Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(1): 12-23. |
| [4] | LIU Yu-ling, WANG Meng-yao, SUN Qi, MA Li-hua, ZHU Xin-xia. Effect of RD29A Promoter on the Stress Resistance of Transgenic Tobacco with SikCDPK1 Gene from Saussurea involucrata [J]. Biotechnology Bulletin, 2023, 39(9): 168-175. |
| [5] | WANG Bao-bao, WANG Hai-yang. Molecular Design of Ideal Plant Architecture for High-density Tolerance of Maize Plant [J]. Biotechnology Bulletin, 2023, 39(8): 11-30. |
| [6] | JIANG Run-hai, JIANG Ran-ran, ZHU Cheng-qiang, HOU Xiu-li. Research Progress in Mechanisms of Microbial-enhanced Phytoremediation for Lead-contaminated Soil [J]. Biotechnology Bulletin, 2023, 39(8): 114-125. |
| [7] | HAN Zhi-yang, JIA Zi-miao, LIANG Qiu-ju, WANG Ke, TANG Hua-li, YE Xing-guo, ZHANG Shuang-xi. Salt Tolerance at Seedling Stage and Analysis of Selenium and Folic Acid Content in Seeds in Two Sets of Wheat-Dasypyrum villosum Chromosom Additional Lines [J]. Biotechnology Bulletin, 2023, 39(8): 185-193. |
| [8] | WANG Yu, YIN Ming-shen, YIN Xiao-yan, XI Jia-qin, YANG Jian-wei, NIU Qiu-hong. Screening, Identification and Degradation Characteristics of Nicotine-degrading Bacteria in Lasioderma serricorne [J]. Biotechnology Bulletin, 2023, 39(6): 308-315. |
| [9] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [10] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [11] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [12] | WANG Feng-ting, WANG Yan, SUN Ying, CUI Wen-jing, QIAO Kai-bin, PAN Hong-yu, LIU Jin-liang. Isolation and Identification of Saline-alkali Tolerant Aspergillus terreus SYAT-1 and Its Activities Against Plant Pathogenic Fungi [J]. Biotechnology Bulletin, 2023, 39(2): 203-210. |
| [13] | LU Zhen-wan, LI Xue-qi, HUANG Jin-guang, ZHOU Huan-bin. Creation of Glyphosate-tolerant Rice by Cytosine Base Editing [J]. Biotechnology Bulletin, 2023, 39(2): 63-69. |
| [14] | CHEN Yi-bo, YANG Wan-ming, YUE Ai-qin, WANG Li-xiang, DU Wei-jun, WANG Min. Construction of Soybean Genetic Map Based on SLAF Markers and QTL Mapping Analysis of Salt Tolerance at Seedling Stage [J]. Biotechnology Bulletin, 2023, 39(2): 70-79. |
| [15] | YANG Mao, LIN Yu-feng, DAI Yang-shuo, PAN Su-jun, PENG Wei-ye, YAN Ming-xiong, LI Wei, WANG Bing, DAI Liang-ying. OsDIS1 Negatively Regulates Rice Drought Tolerance Through Antioxidant Pathways [J]. Biotechnology Bulletin, 2023, 39(2): 88-95. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||