








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (2): 65-72.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0879
Previous Articles Next Articles
GAO Deng-ke1,2( ), MA Bai-rong1,2, GUO Yi-ying1, LIU Wei1,2, LIU Tian1,2, JIN Ya-ping1,2, JIANG Zhou3(
), MA Bai-rong1,2, GUO Yi-ying1, LIU Wei1,2, LIU Tian1,2, JIN Ya-ping1,2, JIANG Zhou3( ), CHEN Hua-tao1,2(
), CHEN Hua-tao1,2( )
)
Received:2023-09-12
Online:2024-02-26
Published:2024-03-13
Contact:
JIANG Zhou, CHEN Hua-tao
E-mail:gdk960101@nwafu.edu.cn;jweiz@126.com;htchen@nwafu.edu.cn
GAO Deng-ke, MA Bai-rong, GUO Yi-ying, LIU Wei, LIU Tian, JIN Ya-ping, JIANG Zhou, CHEN Hua-tao. Establishment of Quaking Knockout Mouse Embryonic Fibroblast Cell Line Using CRISPR/Cas9 Technology[J]. Biotechnology Bulletin, 2024, 40(2): 65-72.
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| sgRNA1 | CACCGAAGTGCAGAATTGCCTGACG |
| AAACCGTCAGGCAATTCTGCACTTC | |
| sgRNA2 | CACCGGATCTTCAACCACCTCGAG |
| AAACCTCGAGGTGGTTGAAGATCC | |
| Target-F | TTATTTCCCAAAGTGAC |
| Target-R | ATGCCCGAATAGGTT |
Table 1 Sequences information
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| sgRNA1 | CACCGAAGTGCAGAATTGCCTGACG |
| AAACCGTCAGGCAATTCTGCACTTC | |
| sgRNA2 | CACCGGATCTTCAACCACCTCGAG |
| AAACCTCGAGGTGGTTGAAGATCC | |
| Target-F | TTATTTCCCAAAGTGAC |
| Target-R | ATGCCCGAATAGGTT |
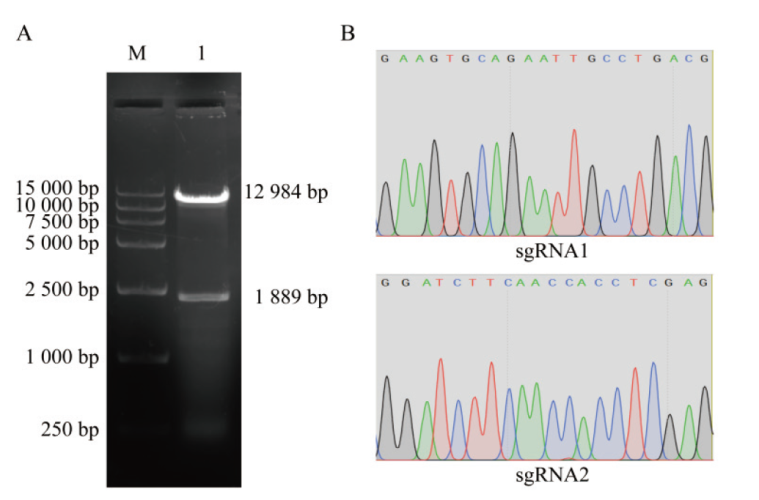
Fig. 2 Construction and identification of recombinant plasmids A: Electrophoresis of LentiCRISPRv2 empty vector enzyme digestion, M: marker; 1: empty carrier of LentiCRISPRv2 after digestion by Esp3I. B: Sequencing results of recombinant plasmids LentiCRISPRv2-sgRNA1 and LentiCRISPRv2-sgRNA2
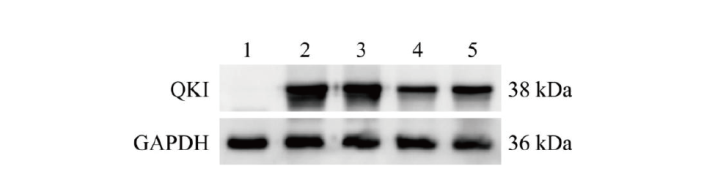
Fig. 3 Screening of recombinant plasmids with high QKI protein knockout efficiency via Western blot 1: Blank control cells without transfection; 2: transfected pcDNA3.1-Quaking overexpression vector; 3: co-transfected pcDNA3.1-Quaking and LentiCRISPRv2 empty vectors; 4: co-transfected pcDNA3.1-Quaking and LentiCRISPRv2-sgRNA1; 5: co-transfected pcDNA3.1-Quaking and LentiCRISPRv2-sgRNA2

Fig. 4 Schematic diagram of comparison between sequencing results of Quaking knockout cell lines and wild-type cell lines Wild-type: Quaking wild-type cell line sequence; Quaking KO: Quaking knockout cell line sequence; sgRNAs are marked in black, PAM sequences in red, dashed blue lines indicate the number of bases removed, and numbers in parentheses indicate the number of bases removed
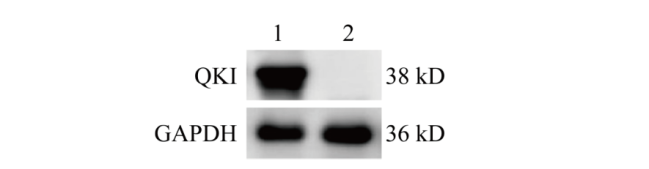
Fig. 6 Detection of QKI protein expression via Western blot 1: NIH3T3 cell samples in control group; 2: NIH3T3 cell samples knocked out by Quaking gene
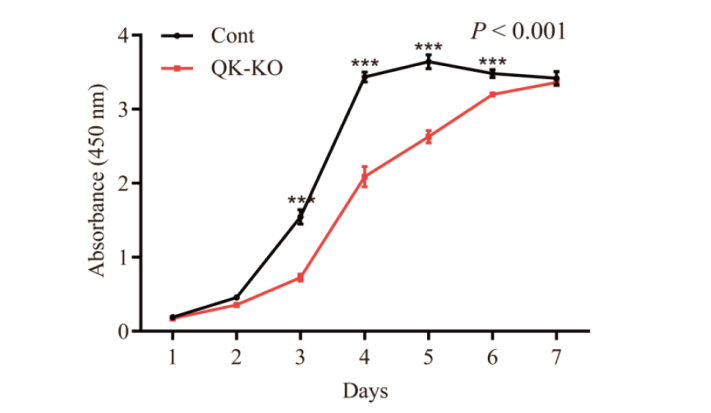
Fig. 8 Effects of Quaking gene knockout on the proliferation ability of NIH3T3 cells P < 0.001 indicates that the cell proliferation activity was significantly different between the two groups in two-factor ANOVA; and ***P < 0.001 indicates a very significant difference
| [1] |
Biedermann B, Hotz HR, Ciosk R. The Quaking family of RNA-binding proteins: coordinators of the cell cycle and differentiation[J]. Cell Cycle, 2010, 9(10): 1929-1933.
pmid: 20495365 |
| [2] |
Ebersole TA, Chen Q, Justice MJ, et al. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins[J]. Nat Genet, 1996, 12(3): 260-265.
doi: 10.1038/ng0396-260 pmid: 8589716 |
| [3] |
Wang YL, Vogel G, Yu ZB, et al. The QKI-5 and QKI-6 RNA binding proteins regulate the expression of microRNA 7 in glial cells[J]. Mol Cell Biol, 2013, 33(6): 1233-1243.
doi: 10.1128/MCB.01604-12 pmid: 23319046 |
| [4] |
Suiko T, Kobayashi K, Aono K, et al. Expression of quaking RNA-binding protein in the adult and developing mouse retina[J]. PLoS One, 2016, 11(5): e0156033.
doi: 10.1371/journal.pone.0156033 URL |
| [5] |
Zhao Y, Zhang G, Wei MY, et al. The tumor suppressing effects of QKI-5 in prostate cancer: a novel diagnostic and prognostic protein[J]. Cancer Biol Ther, 2014, 15(1): 108-118.
doi: 10.4161/cbt.26722 pmid: 24153116 |
| [6] | 赵易, 赵庆丽, 马骥, 等. RNA结合蛋白QKI-5对乳腺癌细胞MCF-7增殖的影响研究[J]. 现代生物医学进展, 2017, 17(25): 4816-4819. |
| Zhao Y, Zhao QL, Ma J, et al. Effect of RNA binding protein of QKI-5 on breast cancer cell MCF-7 of proliferation[J]. Prog Mod Biomed, 2017, 17(25): 4816-4819. | |
| [7] | 段小霞, 郭华, 付婷, 等. 震颤同系物-5过表达对卵巢癌细胞A2780增殖和凋亡的影响[J]. 陕西医学杂志, 2022, 51(5): 529-533. |
| Duan XX, Guo H, Fu T, et al. Effect of quaking homolog-5 overexpression on proliferation and apoptosis of ovarian cancer cell line A2780[J]. Shaanxi Med J, 2022, 51(5): 529-533. | |
| [8] |
Wang L, Zhai DS, Ruan BJ, et al. Quaking deficiency amplifies inflammation in experimental endotoxemia via the aryl hydrocarbon receptor/signal transducer and activator of transcription 1-NF-κB pathway[J]. Front Immunol, 2017, 8: 1754.
doi: 10.3389/fimmu.2017.01754 pmid: 29276519 |
| [9] |
Hardy RJ, Loushin CL, Friedrich VL Jr, et al. Neural cell type-specific expression of QKI proteins is altered in quakingviable mutant mice[J]. J Neurosci, 1996, 16(24): 7941-7949.
pmid: 8987822 |
| [10] |
Larocque D, Pilotte J, Chen TP, et al. Nuclear retention of MBP mRNAs in the quaking viable mice[J]. Neuron, 2002, 36(5): 815-829.
doi: 10.1016/s0896-6273(02)01055-3 pmid: 12467586 |
| [11] |
Noveroske JK, Lai LH, Gaussin V, et al. Quaking is essential for blood vessel development[J]. Genesis, 2002, 32(3): 218-230.
pmid: 11892011 |
| [12] |
Bohnsack BL, Lai LH, Northrop JL, et al. Visceral endoderm function is regulated by quaking and required for vascular development[J]. Genesis, 2006, 44(2): 93-104.
pmid: 16470614 |
| [13] | 张杰. NF-κB信号通路对RNA结合蛋白QKI的转录调控及其在骨骼肌中的功能研究[D]. 西安: 第四军医大学, 2009. |
| Zhang J. Transcriptional regulation of RNA binding protein QKI by NF-κB and elucidation of its function in skeletal muscle[D]. Xi'an: The Fourth Military Medical University, 2009. | |
| [14] | 王逢博, 高登科, 李超, 等. 小鼠RNA结合蛋白QKI-5的生物信息学和组织表达分析及其过表达对癌症相关基因表达的影响[J]. 江西农业大学学报, 2023, 45(2): 453-466. |
| Wang FB, Gao DK, Li C, et al. Bioinformatics and tissue expression analysis of mouse RNA binding protein QKI-5 and its overexpression effect on cancer-related genes expression[J]. Acta Agric Univ Jiangxiensis, 2023, 45(2): 453-466. | |
| [15] |
Gao DK, Ma TT, Gao L, et al. Autophagy activation attenuates the circadian clock oscillators in U2OS cells via the ATG5 pathway[J]. Cell Signal, 2023, 101: 110502.
doi: 10.1016/j.cellsig.2022.110502 URL |
| [16] | 刘思远, 易国强, 唐中林, 等. 基于CRISPR/Cas9系统在全基因组范围内筛选功能基因及调控元件研究进展[J]. 遗传, 2020, 42(5): 435-443. |
| Liu SY, Yi GQ, Tang ZL, et al. Progress on genome-wide CRISPR/Cas9 screening for functional genes and regulatory elements[J]. Hereditas, 2020, 42(5): 435-443. | |
| [17] |
Cong L, Ran FA, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823.
doi: 10.1126/science.1231143 pmid: 23287718 |
| [18] |
Mali P, Yang LH, et al. RNA-guided human genome engineering via Cas9[J]. Science, 2013, 339(6121): 823-826.
doi: 10.1126/science.1232033 pmid: 23287722 |
| [19] |
Jinek M, East A, Cheng A, et al. RNA-programmed genome editing in human cells[J]. eLife, 2013, 2: e00471.
doi: 10.7554/eLife.00471 URL |
| [20] |
Hille F, Richter H, Wong SP, et al. The biology of CRISPR-cas: backward and forward[J]. Cell, 2018, 172(6): 1239-1259.
doi: S0092-8674(17)31383-1 pmid: 29522745 |
| [21] |
Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-cas13[J]. Science, 2017, 358(6366): 1019-1027.
doi: 10.1126/science.aaq0180 pmid: 29070703 |
| [22] |
Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096.
doi: 10.1126/science.1258096 URL |
| [23] |
Kambol R, Gatseva A, Gifford RJ. An endogenous lentivirus in the germline of a rodent[J]. Retrovirology, 2022, 19(1): 30.
doi: 10.1186/s12977-022-00615-2 pmid: 36539757 |
| [24] |
Cockrell AS, Kafri T. Gene delivery by lentivirus vectors[J]. Mol Biotechnol, 2007, 36(3): 184-204.
doi: 10.1007/s12033-007-0010-8 pmid: 17873406 |
| [25] |
You LT, Tong RZ, Li MQ, et al. Advancements and obstacles of CRISPR-Cas9 technology in translational research[J]. Mol Ther Methods Clin Dev, 2019, 13: 359-370.
doi: 10.1016/j.omtm.2019.02.008 URL |
| [26] |
Wang HX, Li MQ, Lee CM, et al. CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery[J]. Chem Rev, 2017, 117(15): 9874-9906.
doi: 10.1021/acs.chemrev.6b00799 URL |
| [27] |
Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases[J]. Nat Rev Genet, 2016, 17(5): 300-312.
doi: 10.1038/nrg.2016.28 pmid: 27087594 |
| [28] |
Wang JZ, Fu X, Fang ZY, et al. QKI-5 regulates the alternative splicing of cytoskeletal gene ADD3 in lung cancer[J]. J Mol Cell Biol, 2021, 13(5): 347-360.
doi: 10.1093/jmcb/mjaa063 URL |
| [29] | 梁梦迪. QKI对猪睾丸支持细胞增殖与分泌因子影响的研究[D]. 长春: 吉林大学, 2019. |
| Liang MD. Effects of QKI on the proliferation and secretion factors of pig Sertoli cells[D]. Changchun: Jilin University, 2019. | |
| [30] |
Chen SJ, Niu S, Wang WN, et al. Overexpression of the QKI gene promotes differentiation of goat myoblasts into myotubes[J]. Animals, 2023, 13(4): 725.
doi: 10.3390/ani13040725 URL |
| [1] | ZHU Tian-yi, KONG Gui-mei, JIAO Hong-mei, GUO Ting-ting, WU Ri-han, LIU Cui-cui, GAO Cheng-feng, LI Guo-cai. Establishment of A Bacterial Model of CRISPR/Cas9 Mediated adeG Gene Knockout in Escherichia coli [J]. Biotechnology Bulletin, 2024, 40(2): 55-64. |
| [2] | ZHANG Hong-min, LONG Wen, LAO Xiao-qing, CHEN Wen-yan, SHANG Xue-mei, WANG Hong-lian, WANG Li, SU Hong-wei, SHEN Hong-ping, SHEN Hong-chun. Construction of Pmepa1 Knockout TCMK1 Mouse Renal Tubular Epithelial Cell Line Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2024, 40(2): 73-79. |
| [3] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [4] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [5] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [6] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [7] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [8] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [9] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [10] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [11] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [12] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [13] | Olalekan Amoo, HU Li-min, ZHAI Yun-gu, FAN Chu-chuan, ZHOU Yong-ming. Regulation of Shoot Branching by BRANCHED1 in Brassica napus Based on Gene Editing Technology [J]. Biotechnology Bulletin, 2022, 38(4): 97-105. |
| [14] | DING Ya-qun, DING Ning, XIE Shen-min, HUANG Meng-na, ZHANG Yu, ZHANG Qin, JIANG Li. Construction of Vps28 Knock-out Mice and Model Study of the Impact on Lactation and Immune Traits [J]. Biotechnology Bulletin, 2022, 38(3): 164-172. |
| [15] | YAN Jiong, FENG Chen-yi, GAO Xue-kun, XU Xiang, YANG Jia-min, CHEN Zhao-yang. Construction of Homozygous Plin1-knockout Mouse Model and Phenotype Analysis Based on CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2022, 38(3): 173-180. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||