








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 199-211.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0139
Previous Articles Next Articles
WANG Qian( ), ZHOU Jia-yan, WANG Qian, DENG Yu-ping, ZHANG Min-hui, CHEN Jing, YANG Jun, ZOU Jian(
), ZHOU Jia-yan, WANG Qian, DENG Yu-ping, ZHANG Min-hui, CHEN Jing, YANG Jun, ZOU Jian( )
)
Received:2024-02-04
Online:2024-08-26
Published:2024-06-26
Contact:
ZOU Jian
E-mail:1347404972@qq.com;zoujian@cwnu.edu.cn
WANG Qian, ZHOU Jia-yan, WANG Qian, DENG Yu-ping, ZHANG Min-hui, CHEN Jing, YANG Jun, ZOU Jian. Identification and Expression Analysis of the YABBY Gene Family in Sunflower[J]. Biotechnology Bulletin, 2024, 40(8): 199-211.
| 基因 Gene | 基因编号 Gene ID | 染色体分布Chromosome distribution | 氨基酸数目 Number of amino acids | 等电点 pI | 分子量 Molecular weight/Da | 亚细胞定位 Subcellular location | ||
|---|---|---|---|---|---|---|---|---|
| 编号No. | 开始位置Starting | 结束位置Ending | ||||||
| HaYABBY 1a | HanXRQr2_Chr06g0245541 | 6 | 12 754 239 | 12 758 093 | 219 | 7.21 | 24 420.73 | 细胞核 |
| HaYABBY 1b | HanXRQr2_Chr13g0613121 | 13 | 161 766 031 | 161 771 918 | 216 | 6.72 | 24 136.35 | 细胞核 |
| HaYABBY 1c | HanXRQr2_Chr04g0185291 | 4 | 195 371 365 | 195 376 174 | 219 | 7.21 | 24 437.79 | 细胞核 |
| HaCRABS CLAWa | HanXRQr2_Chr12g0549931 | 1 | 81 233 913 | 81 243 999 | 173 | 9.4 | 19 366.25 | 细胞核 |
| HaCRABS CLAWb | HanXRQr2_Chr15g0688211 | 15 | 39 093 254 | 39 095 798 | 180 | 9.28 | 19 765.53 | 细胞核 |
| HaYABBY 3 | HanXRQr2_Chr13g0577751 | 13 | 57 727 478 | 57 739 518 | 225 | 9.16 | 24 724.41 | 细胞核 |
| HaYABBY 4a | HanXRQr2_Chr17g0787111 | 17 | 15 387 218 | 15 388 714 | 197 | 4.98 | 21 878.58 | 细胞核 |
| HaYABBY 4b | HanXRQr2_Chr06g0253091 | 6 | 30 699 177 | 3 700 575 | 236 | 6.4 | 26 603.38 | 细胞核 |
| HaYABBY 4c | HanXRQr2_Chr03g0102331 | 3 | 78 930 712 | 78 931 672 | 99 | 7.82 | 10 936.49 | 细胞核 |
| HaYABBY 5a | HanXRQr2_Chr17g0801501 | 17 | 53 868 011 | 53 872 537 | 227 | 8.55 | 25 164.39 | 细胞核 |
| HaYABBY 5b | HanXRQr2_Chr10g0462601 | 10 | 175 418 675 | 175 424 195 | 200 | 9.74 | 22 555.59 | 细胞核 |
| HaYABBY 5c | HanXRQr2_Chr12g0523971 | 12 | 5 127 939 | 5 135 711 | 186 | 9.67 | 21 053.74 | 细胞核 |
| HaYABBY 5d | HanXRQr2_Chr06g0238981 | 6 | 1 174 948 | 1 181 807 | 229 | 8.5 | 25 139.60 | 细胞核 |
Table 1 Information of YABBY gene family members in sunflower
| 基因 Gene | 基因编号 Gene ID | 染色体分布Chromosome distribution | 氨基酸数目 Number of amino acids | 等电点 pI | 分子量 Molecular weight/Da | 亚细胞定位 Subcellular location | ||
|---|---|---|---|---|---|---|---|---|
| 编号No. | 开始位置Starting | 结束位置Ending | ||||||
| HaYABBY 1a | HanXRQr2_Chr06g0245541 | 6 | 12 754 239 | 12 758 093 | 219 | 7.21 | 24 420.73 | 细胞核 |
| HaYABBY 1b | HanXRQr2_Chr13g0613121 | 13 | 161 766 031 | 161 771 918 | 216 | 6.72 | 24 136.35 | 细胞核 |
| HaYABBY 1c | HanXRQr2_Chr04g0185291 | 4 | 195 371 365 | 195 376 174 | 219 | 7.21 | 24 437.79 | 细胞核 |
| HaCRABS CLAWa | HanXRQr2_Chr12g0549931 | 1 | 81 233 913 | 81 243 999 | 173 | 9.4 | 19 366.25 | 细胞核 |
| HaCRABS CLAWb | HanXRQr2_Chr15g0688211 | 15 | 39 093 254 | 39 095 798 | 180 | 9.28 | 19 765.53 | 细胞核 |
| HaYABBY 3 | HanXRQr2_Chr13g0577751 | 13 | 57 727 478 | 57 739 518 | 225 | 9.16 | 24 724.41 | 细胞核 |
| HaYABBY 4a | HanXRQr2_Chr17g0787111 | 17 | 15 387 218 | 15 388 714 | 197 | 4.98 | 21 878.58 | 细胞核 |
| HaYABBY 4b | HanXRQr2_Chr06g0253091 | 6 | 30 699 177 | 3 700 575 | 236 | 6.4 | 26 603.38 | 细胞核 |
| HaYABBY 4c | HanXRQr2_Chr03g0102331 | 3 | 78 930 712 | 78 931 672 | 99 | 7.82 | 10 936.49 | 细胞核 |
| HaYABBY 5a | HanXRQr2_Chr17g0801501 | 17 | 53 868 011 | 53 872 537 | 227 | 8.55 | 25 164.39 | 细胞核 |
| HaYABBY 5b | HanXRQr2_Chr10g0462601 | 10 | 175 418 675 | 175 424 195 | 200 | 9.74 | 22 555.59 | 细胞核 |
| HaYABBY 5c | HanXRQr2_Chr12g0523971 | 12 | 5 127 939 | 5 135 711 | 186 | 9.67 | 21 053.74 | 细胞核 |
| HaYABBY 5d | HanXRQr2_Chr06g0238981 | 6 | 1 174 948 | 1 181 807 | 229 | 8.5 | 25 139.60 | 细胞核 |
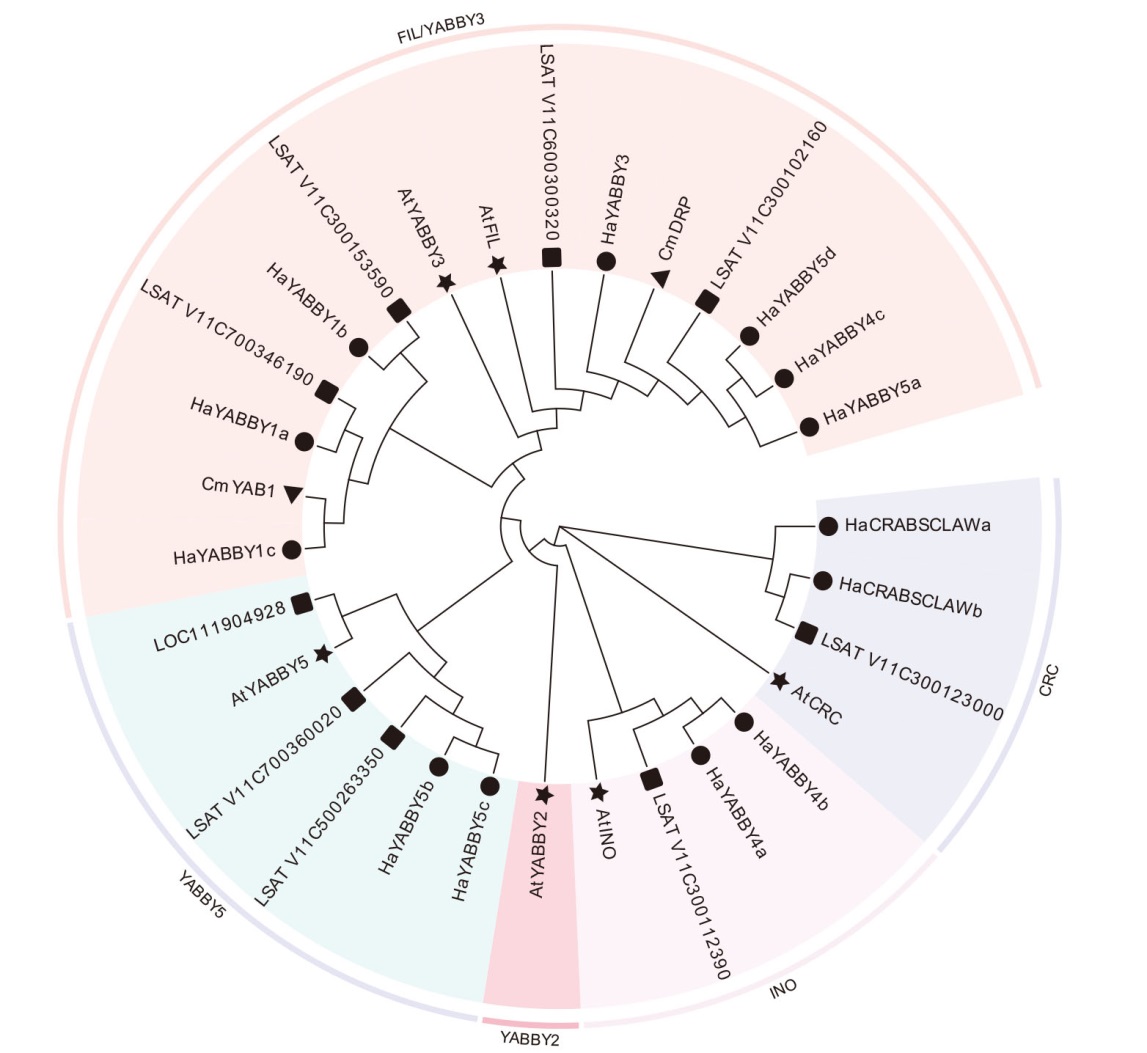
Fig. 2 Phylogenetic tree analysis of YABBY gene family members in sunflower ★,●,▲ and ■indicate Arabidopsis, sunflower, Chrysanthemum morifolium and Lactuca sativa respectively

Fig. 3 Gene structure and conserved motifs for YABBY family members A: Mapping of conserved motifs for YABBY family members. B: Mapping of gene structure for YABBY family members. C: Amino acid sequence comparison of core conserved domain of YABBY family members
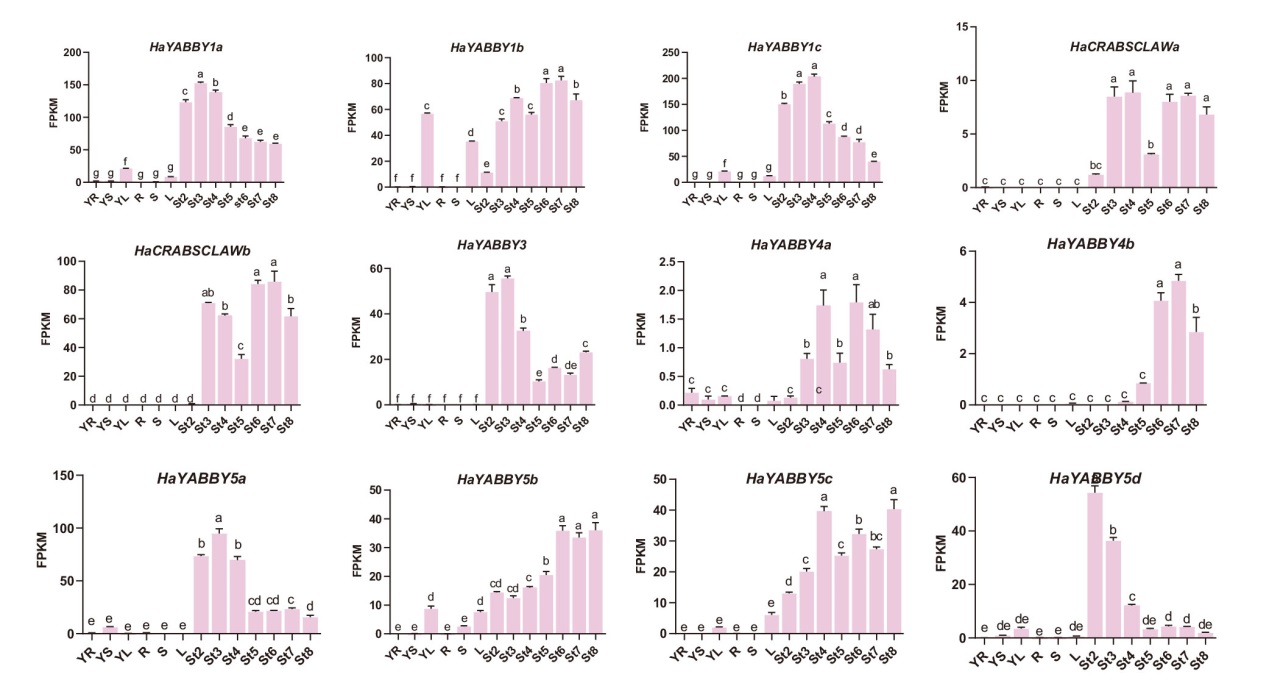
Fig. 4 Analysis of tissue expression pattern of YABBY gene in WT sunflower FPKM refers to transcriptional abundance of YABBY gene family. Tissues: YR: young root; YS: young stem; YL: young leaf; R: root; S: stem; L: leaf. Expression period: St2: flower primordium initiation stage; St3: flower organ formation stage; St4: meiosis stage; St5: pollen maturation stage; St6: florets opening stage; St7: fruit filling stage and St8 seed full stage. The lowercase letter indicates significant difference at P<0.05 level. The same below

Fig. 5 Differential expression analysis of YABBY gene in WT sunflower and cb1 mutant A: Phenotypic characteristics of WT sunflower and cb1 mutant inflorescence. B: Phenotypic characteristics of inflorescences in WT and cb1 mutant sunflower from St2-St8. C: Relative expression of YABBY gene in WT and cb1 during inflorescence development(st2-st8). All data are mean ±SD(n=3). *: Significant difference(P<0.05); **: extremely significant difference(P<0.01)
| [1] |
Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development[J]. Trends Genet, 2010, 26(12): 519-527.
doi: 10.1016/j.tig.2010.09.001 pmid: 20947199 |
| [2] |
Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, et al. DNA-binding specificities of plant transcription factors and their potential to define target genes[J]. Proc Natl Acad Sci USA, 2014, 111(6): 2367-2372.
doi: 10.1073/pnas.1316278111 pmid: 24477691 |
| [3] |
Porto MS, Pinheiro MPN, Batista VGL, et al. Plant promoters: an approach of structure and function[J]. Mol Biotechnol, 2014, 56(1): 38-49.
pmid: 24122284 |
| [4] |
Udvardi MK, Kakar K, Wandrey M, et al. Legume transcription factors: global regulators of plant development and response to the environment[J]. Plant Physiol, 2007, 144(2): 538-549.
doi: 10.1104/pp.107.098061 pmid: 17556517 |
| [5] | 刁志娟. 水稻三个花器官发育关键基因和一个SUPERMAN-like基因的功能和表达分析[D]. 福州: 福建农林大学, 2011. |
| Diao ZJ. Functional and expressional analysis of three pivotal genes for floral organ development and a SUPERMAN-like gene in rice(Oryza sativa L.)[D]. Fuzhou: Fujian Agriculture and Forestry University, 2011. | |
| [6] | Nam J, DePamphilis CW, Ma H, et al. Antiquity and evolution of the MADS-box gene family controlling flower development in plants[J]. Mol Biol Evol, 2003, 20(9): 1435-1447. |
| [7] | Mantegazza O, Gregis V, Mendes MA, et al. Analysis of the Arabidopsis REM gene family predicts functions during flower development[J]. Ann Bot, 2014, 114(7): 1507-1515. |
| [8] | Costanzo E, Trehin C, Vandenbussche M. The role of WOX genes in flower development[J]. Ann Bot, 2014, 114(7): 1545-1553. |
| [9] | Viola IL, Gonzalez DH. TCP transcription factors in plant reproductive development: juggling multiple roles[J]. Biomolecules, 2023, 13(5): 750. |
| [10] | Yamada T, Yokota S, Hirayama Y, et al. Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms[J]. Plant J, 2011, 67(1): 26-36. |
| [11] | Bartholmes C, Hidalgo O, Gleissberg S. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica(Papaveraceae)[J]. Plant Biol, 2012, 14(1): 11-23. |
| [12] | Finet C, Floyd SK, Conway SJ, et al. Evolution of the YABBY gene family in seed plants[J]. Evol Dev, 2016, 18(2): 116-126. |
| [13] |
Zhang TP, Li CY, Li DX, et al. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants[J]. J Plant Res, 2020, 133(6): 751-763.
doi: 10.1007/s10265-020-01227-7 pmid: 33033876 |
| [14] | Romanova MA, Maksimova AI, Pawlowski K, et al. YABBY genes in the development and evolution of land plants[J]. Int J Mol Sci, 2021, 22(8): 4139. |
| [15] |
Siegfried KR, Eshed Y, Baum SF, et al. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis[J]. Development, 1999, 126(18): 4117-4128.
doi: 10.1242/dev.126.18.4117 pmid: 10457020 |
| [16] | Sawa S, Watanabe K, Goto K, et al. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains[J]. Genes Dev, 1999, 13(9): 1079-1088. |
| [17] |
Bowman JL. The YABBY gene family and abaxial cell fate[J]. Curr Opin Plant Biol, 2000, 3(1): 17-22.
doi: 10.1016/s1369-5266(99)00035-7 pmid: 10679447 |
| [18] | Lee JY, Baum SF, Oh SH, et al. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade[J]. Development, 2005, 132(22): 5021-5032. |
| [19] | Yamada T, Ito M, Kato M. YABBY2-homologue expression in lateral organs of Amborella trichopoda(Amborellaceae)[J]. Int J Plant Sci, 2004, 165(6): 917-924. |
| [20] |
Sawa S, Ito T, Shimura Y, et al. FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems[J]. Plant Cell, 1999, 11(1): 69-86.
pmid: 9878633 |
| [21] |
Bowman JL, Smyth DR. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains[J]. Development, 1999, 126(11): 2387-2396.
doi: 10.1242/dev.126.11.2387 pmid: 10225998 |
| [22] | Villanueva JM, Broadhvest J, Hauser BA, et al. INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules[J]. Genes Dev, 1999, 13(23): 3160-3169. |
| [23] | Eshed Y, Izhaki A, Baum SF, et al. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities[J]. Development, 2004, 131(12): 2997-3006. |
| [24] | Sarojam R, Sappl PG, Goldshmidt A, et al. Differentiating Arabidopsis shoots from leaves by combined YABBY activities[J]. Plant Cell, 2010, 22(7): 2113-2130. |
| [25] |
Chen Q, Atkinson A, Otsuga D, et al. The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation[J]. Development, 1999, 126(12): 2715-2726.
doi: 10.1242/dev.126.12.2715 pmid: 10331982 |
| [26] |
Lugassi N, Nakayama N, Bochnik R, et al. A novel allele of FILAMENTOUS FLOWER reveals new insights on the link between inflorescence and floral meristem organization and flower morphogenesis[J]. BMC Plant Biol, 2010, 10: 131.
doi: 10.1186/1471-2229-10-131 pmid: 20584289 |
| [27] | Fourquin C, Vinauger-Douard M, Fogliani B, et al. Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms[J]. Proc Natl Acad Sci U S A, 2005, 102(12): 4649-4654. |
| [28] |
Yamaguchi T, Nagasawa N, Kawasaki S, et al. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa[J]. Plant Cell, 2004, 16(2): 500-509.
doi: 10.1105/tpc.018044 pmid: 14729915 |
| [29] |
Ishikawa M, Ohmori Y, Tanaka W, et al. The spatial expression patterns of DROOPING LEAF orthologs suggest a conserved function in grasses[J]. Genes Genet Syst, 2009, 84(2): 137-146.
pmid: 19556707 |
| [30] | Orashakova S, Lange M, Lange S, et al. The CRABS CLAW ortholog from California poppy(Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation[J]. Plant J, 2009, 58(4): 682-693. |
| [31] |
Balasubramanian S, Schneitz K. NOZZLE links proximal-distal and adaxial-abaxial pattern formation during ovule development in Arabidopsis thaliana[J]. Development, 2002, 129(18): 4291-4300.
doi: 10.1242/dev.129.18.4291 pmid: 12183381 |
| [32] |
Meister RJ, Kotow LM, Gasser CS. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments[J]. Development, 2002, 129(18): 4281-4289.
doi: 10.1242/dev.129.18.4281 pmid: 12183380 |
| [33] | di Rienzo V, Imanifard Z, Mascio I, et al. Functional conservation of the grapevine candidate gene INNER NO OUTER for ovule development and seed formation[J]. Hortic Res, 2021, 8(1): 29. |
| [34] | Tanaka W, Toriba T, Ohmori Y, et al. The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet[J]. Plant Cell, 2012, 24(1): 80-95. |
| [35] | Ali Buttar Z, Yang Y, Sharif R, et al. Genome wide identification, characterization, and expression analysis of YABBY-gene family in wheat (Triticum aestivum L.)[J]. Agronomy, 2020, 10(8): 1189. |
| [36] | Chen YY, Hsiao YY, Chang SB, et al. Genome-wide identification of YABBY genes in Orchidaceae and their expression patterns in Phalaenopsis orchid[J]. Genes, 2020, 11(9): 955. |
| [37] |
Cong B, Barrero LS, Tanksley SD. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication[J]. Nat Genet, 2008, 40(6): 800-804.
doi: 10.1038/ng.144 pmid: 18469814 |
| [38] |
Li Z, Zhang ZH, Yan PC, et al. RNA-Seq improves annotation of protein-coding genes in the cucumber genome[J]. BMC Genomics, 2011, 12: 540.
doi: 10.1186/1471-2164-12-540 pmid: 22047402 |
| [39] |
Zhang X, Ding L, Song AP, et al. DWARF AND ROBUST PLANT regulates plant height via modulating gibberellin biosynthesis in chrysanthemum[J]. Plant Physiol, 2022, 190(4): 2484-2500.
doi: 10.1093/plphys/kiac437 pmid: 36214637 |
| [40] |
Gross T, Broholm S, Becker A. CRABS CLAW acts as a bifunctional transcription factor in flower development[J]. Front Plant Sci, 2018, 9: 835.
doi: 10.3389/fpls.2018.00835 pmid: 29973943 |
| [41] | Yang ZE, Gong Q, Wang LL, et al. Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses[J]. Front Genet, 2018, 9: 33. |
| [42] | Wils CR, Kaufmann K. Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana[J]. Biochim Biophys Acta Gene Regul Mech, 2017, 1860(1): 95-105. |
| [43] | Lee DY, An G. Two AP2 family genes, SUPERNUMERARY BRACT(SNB)and OsINDETERMINATE SPIKELET 1(OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice[J]. Plant J, 2012, 69(3): 445-461. |
| [44] | Melzer S, Lens F, Gennen J, et al. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana[J]. Nat Genet, 2008, 40(12): 1489-1492. |
| [45] | Jiu ST, Zhang YP, Han P, et al. Genome-wide identification and expression analysis of VviYABs family reveal its potential functions in the developmental switch and stresses response during grapevine development[J]. Front Genet, 2022, 12: 762221. |
| [46] | Mäkilä R, Wybouw B, Smetana O, et al. Gibberellins promote polar auxin transport to regulate stem cell fate decisions in cambium[J]. Nat Plants, 2023, 9(4): 631-644. |
| [47] | Freire-Rios A, Tanaka K, Crespo I, et al. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis[J]. Proc Natl Acad Sci U S A, 2020, 117(39): 24557-24566. |
| [48] |
Vaddepalli P, Scholz S, Schneitz K. Pattern formation during early floral development[J]. Curr Opin Genet Dev, 2015, 32: 16-23.
doi: 10.1016/j.gde.2015.01.001 pmid: 25687790 |
| [49] | Dai MQ, Hu YF, Zhao Y, et al. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development[J]. Plant Physiol, 2007, 144(1): 380-390. |
| [50] | Siefers N, Dang KK, Kumimoto RW, et al. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity[J]. Plant Physiol, 2009, 149(2): 625-641. |
| [1] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [2] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [3] | QIN Jian, LI Zhen-yue, HE Lang, LI Jun-ling, ZHANG Hao, DU Rong. Change of Single-cell Transcription Profile and Analysis of Intercellular Communication in Myogenic Cell Differentiation [J]. Biotechnology Bulletin, 2024, 40(6): 330-342. |
| [4] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [5] | CHEN Qiang, HUANG Xin-hui, ZHANG Zheng, ZHANG Chong, LIU Ye-fei. Effects of Melatonin on the Fruit Softening and Ethylene Synthesis of Post-harvest Oriental Melon [J]. Biotechnology Bulletin, 2024, 40(4): 139-147. |
| [6] | ZHANG Yu, SHI Lei, GONG Lei, NIE Feng-jie, YANG Jiang-wei, LIU Xuan, YANG Wen-jing, ZHANG Guo-hui, XIE Rui-xia, ZHANG Li. Genome-wide Identification of Potato WOX Gene Family and Its Expression Analysis in in vitro Regeneration and Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(3): 170-180. |
| [7] | WU Xing-xing, HONG Hai-bo, GAN Zhi-cheng, LI Rui-ning, HUANG Xian-zhong. Cloning and Preliminary Functional Analysis of CaPI Gene in Capsicum annuum L. [J]. Biotechnology Bulletin, 2024, 40(3): 193-201. |
| [8] | JIANG Lin-qi, ZHAO Jia-ying, ZHENG Fei-xiong, YAO Xin-yi, LI Xiao-xian, YU Zhen-ming. Identification and Expression Analysis of 14-3-3 Gene Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2024, 40(3): 229-241. |
| [9] | WU Zhen, ZHANG Ming-Ying, YAN Feng, LI Yi-min, GAO Jing, YAN Yong-Gang, ZHANG Gang. Identification and Analysis of WRKY Gene Family in Rheum palmatum L. [J]. Biotechnology Bulletin, 2024, 40(1): 250-261. |
| [10] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [11] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [12] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [13] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [14] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [15] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||