








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (9): 148-160.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1176
Previous Articles Next Articles
MA Bo-tao( ), WU Guo-qiang(
), WU Guo-qiang( ), WEI Ming
), WEI Ming
Received:2023-12-13
Online:2024-09-26
Published:2024-10-12
Contact:
WU Guo-qiang
E-mail:2782130164@qq.com;gqwu@lut.edu.cn
MA Bo-tao, WU Guo-qiang, WEI Ming. Roles of bZIP Transcription Factor in the Response to Stresses, and Growth and Development in Plants[J]. Biotechnology Bulletin, 2024, 40(9): 148-160.
| 物种Species | 基因名称Gene name | 基因数目Number of genes | 亚家族数目Number of subfamilies | 参考文献Reference |
|---|---|---|---|---|
| 杨树Populus simonii | PlbZIP | 86 | 12 | [ |
| 拟南芥A. thaliana | AtbZIP | 78 | 13 | [ |
| 木薯M. esculenta | MebZIP | 77 | 10 | [ |
| 菠萝A. comosus | AcbZIP | 57 | 11 | [ |
| 烟草N. tabacum | NtbZIP | 77 | 11 | [ |
| 马铃薯S. tuberosum | StbZIP | 56 | 10 | [ |
| 百香果P. Sims | PebZIP | 56 | 12 | [ |
| 大豆G. max | GmbZIP | 160 | 12 | [ |
| 水稻O. sativa | OsbZIP | 89 | 10 | [ |
| 红花Carthamus tinctorius | CtbZIP | 52 | 12 | [ |
| 草莓Fragaria vesca | FvbZIP | 50 | 20 | [ |
| 小麦Triticum aestivum | TabZIP | 227 | 13 | [ |
| 辣椒Capsicum annuum | CabZIP | 60 | 10 | [ |
| 苹果Malus domestica | MdbZIP | 112 | 11 | [ |
| 甘蓝Brassica oleracea | BobZIP | 119 | 63 | [ |
| 猕猴桃Actinidia chinensis | AlbZIP | 81 | 11 | [ |
| 绿豆Vigna radiata | VrbZIP | 75 | 10 | [ |
| 枣树Zizyphus jujuba | ZjbZIP | 45 | 14 | [ |
| 甜菜Beta vulgaris | BvbZIP | 48 | 6 | [ |
| 番茄Solanum lycopersicum | SlbZIP | 69 | 9 | [ |
| 玉米Zea mays | ZmbZIP | 125 | 11 | [ |
| 大麻Cannabis sativa | CsbZIP | 51 | 11 | [ |
| 核桃Juglans regia | JrbZIP | 88 | 13 | [ |
| 荔枝Litchi chinensis | LcbZIP | 54 | 14 | [ |
| 毛竹Phyllostachys edulis | PhebZIP | 154 | 9 | [ |
| 蓖麻Ricinus communis | RcbZIP | 49 | 11 | [ |
| 苦荞Fagopyum talaricum | FtbZIP | 96 | 11 | [ |
| 人参Panax ginseng | PgbZIP | 91 | 10 | [ |
| 黄瓜Cucumis sativus | CsbZIP | 64 | 6 | [ |
| 葡萄Vitis vinifera | VvbZIP | 55 | 10 | [ |
| 芝麻Sesamum indicum | SibZIP | 63 | 9 | [ |
| 梨Pyrus bretschneideri | PbbZIP | 62 | 13 | [ |
| 桃子Prunus persica | PpbZIP | 50 | 10 | [ |
| 橄榄Olea europaea | OebZIP | 103 | 12 | [ |
| 棉花Gossypium hirsutum | GhbZIP | 197 | 13 | [ |
| 菘蓝Isatis indigotica | IibZIP | 65 | 11 | [ |
| 铁皮石斛Dendrobium catenatum | DcbZIP | 62 | 10 | [ |
| 甘薯Ipomoea batatas | IbbZIP | 87 | 11 | [ |
| 甜橙Citrus sinensis | CsebZIP | 50 | 16 | [ |
Table 1 bZIP genes in different plant species
| 物种Species | 基因名称Gene name | 基因数目Number of genes | 亚家族数目Number of subfamilies | 参考文献Reference |
|---|---|---|---|---|
| 杨树Populus simonii | PlbZIP | 86 | 12 | [ |
| 拟南芥A. thaliana | AtbZIP | 78 | 13 | [ |
| 木薯M. esculenta | MebZIP | 77 | 10 | [ |
| 菠萝A. comosus | AcbZIP | 57 | 11 | [ |
| 烟草N. tabacum | NtbZIP | 77 | 11 | [ |
| 马铃薯S. tuberosum | StbZIP | 56 | 10 | [ |
| 百香果P. Sims | PebZIP | 56 | 12 | [ |
| 大豆G. max | GmbZIP | 160 | 12 | [ |
| 水稻O. sativa | OsbZIP | 89 | 10 | [ |
| 红花Carthamus tinctorius | CtbZIP | 52 | 12 | [ |
| 草莓Fragaria vesca | FvbZIP | 50 | 20 | [ |
| 小麦Triticum aestivum | TabZIP | 227 | 13 | [ |
| 辣椒Capsicum annuum | CabZIP | 60 | 10 | [ |
| 苹果Malus domestica | MdbZIP | 112 | 11 | [ |
| 甘蓝Brassica oleracea | BobZIP | 119 | 63 | [ |
| 猕猴桃Actinidia chinensis | AlbZIP | 81 | 11 | [ |
| 绿豆Vigna radiata | VrbZIP | 75 | 10 | [ |
| 枣树Zizyphus jujuba | ZjbZIP | 45 | 14 | [ |
| 甜菜Beta vulgaris | BvbZIP | 48 | 6 | [ |
| 番茄Solanum lycopersicum | SlbZIP | 69 | 9 | [ |
| 玉米Zea mays | ZmbZIP | 125 | 11 | [ |
| 大麻Cannabis sativa | CsbZIP | 51 | 11 | [ |
| 核桃Juglans regia | JrbZIP | 88 | 13 | [ |
| 荔枝Litchi chinensis | LcbZIP | 54 | 14 | [ |
| 毛竹Phyllostachys edulis | PhebZIP | 154 | 9 | [ |
| 蓖麻Ricinus communis | RcbZIP | 49 | 11 | [ |
| 苦荞Fagopyum talaricum | FtbZIP | 96 | 11 | [ |
| 人参Panax ginseng | PgbZIP | 91 | 10 | [ |
| 黄瓜Cucumis sativus | CsbZIP | 64 | 6 | [ |
| 葡萄Vitis vinifera | VvbZIP | 55 | 10 | [ |
| 芝麻Sesamum indicum | SibZIP | 63 | 9 | [ |
| 梨Pyrus bretschneideri | PbbZIP | 62 | 13 | [ |
| 桃子Prunus persica | PpbZIP | 50 | 10 | [ |
| 橄榄Olea europaea | OebZIP | 103 | 12 | [ |
| 棉花Gossypium hirsutum | GhbZIP | 197 | 13 | [ |
| 菘蓝Isatis indigotica | IibZIP | 65 | 11 | [ |
| 铁皮石斛Dendrobium catenatum | DcbZIP | 62 | 10 | [ |
| 甘薯Ipomoea batatas | IbbZIP | 87 | 11 | [ |
| 甜橙Citrus sinensis | CsebZIP | 50 | 16 | [ |
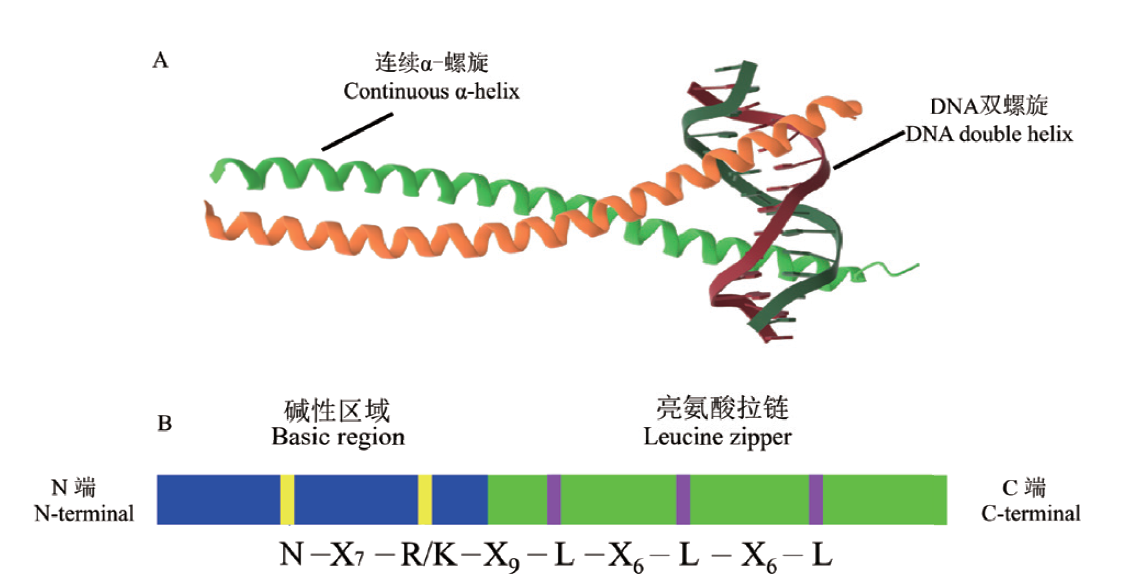
Fig. 1 Model of bZIP structure A: bZIP transcription factor protein model map, is leucine zipper region and DNA-binding basic region respectively, which is derived by PDB website(www.rcsb.org). B: The consistency sequence of bZIP

Fig. 2 Phylogenetic tree of bZIPs in higher plant Clustal W software was used for multiple alignment of sequences, the phylogenetic tree was constructed using MEGA 11 software. The red circle indicates Arabidopsis thaliana, the green stars indicate Beta vulgaris, and the blue triangle indicate Oryza sativa. The name, source, and accession number of bZIPs are shown in the Supplementary Table 1
| [1] | Javed T, Shabbir R, Ali A, et al. Transcription factors in plant stress responses: challenges and potential for sugarcane improvement[J]. Plants, 2020, 9(4): 491. |
| [2] | Waadt R, Seller CA, Hsu PK, et al. Plant hormone regulation of abiotic stress responses[J]. Nat Rev Mol Cell Biol, 2022, 23(10): 680-694. |
| [3] | Zhang Q, Liu YQ, Jiang YL, et al. OsASR6 enhances salt stress tolerance in rice[J]. Int J Mol Sci, 2022, 23(16): 9340. |
| [4] | Guo R, Shi LX, Jiao Y, et al. Metabolic responses to drought stress in the tissues of drought-tolerant and drought-sensitive wheat genotype seedlings[J]. AoB Plants, 2018, 10(2): ply016. |
| [5] | Zuo DD, Ahammed GJ, Guo DL. Plant transcriptional memory and associated mechanism of abiotic stress tolerance[J]. Plant Physiol Biochem, 2023, 201: 107917. |
| [6] | Fortunato S, Lasorella C, Dipierro N, et al. Redox signaling in plant heat stress response[J]. Antioxidants, 2023, 12(3): 605. |
| [7] | Yoon Y, Seo DH, Shin H, et al. The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants[J]. Agronomy, 2020, 10(6): 788. |
| [8] |
Zhao K, Chen S, Yao WJ, et al. Genome-wide analysis and expression profile of the bZIP gene family in poplar[J]. BMC Plant Biol, 2021, 21(1): 122.
doi: 10.1186/s12870-021-02879-w pmid: 33648455 |
| [9] |
Jakoby M, Weisshaar B, Dröge-Laser W, et al. bZIP transcription factors in Arabidopsis[J]. Trends Plant Sci, 2002, 7(3): 106-111.
doi: 10.1016/s1360-1385(01)02223-3 pmid: 11906833 |
| [10] |
Hu W, Yang HB, Yan Y, et al. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava[J]. Sci Rep, 2016, 6: 22783.
doi: 10.1038/srep22783 pmid: 26947924 |
| [11] | Liu YH, Chai MN, Zhang M, et al. Genome-wide analysis, characterization, and expression profile of the basic leucine zipper transcription factor family in pineapple[J]. Int J Genomics, 2020, 2020: 3165958. |
| [12] | Duan LL, Mo ZJ, Fan Y, et al. Genome-wide identification and expression analysis of the bZIP transcription factor family genes in response to abiotic stress in Nicotiana tabacum L[J]. BMC Genomics, 2022, 23(1): 318. |
| [13] | Mirzaei K, Bahramnejad B, Fatemi S. Genome-wide identification and characterization of the bZIP gene family in potato(Solanum tuberosum)[J]. Plant Gene, 2020, 24: 100257. |
| [14] | Ma FN, Zhou HW, Xu Y, et al. Comprehensive analysis of bZIP transcription factors in passion fruit[J]. iScience, 2023, 26(4): 106556. |
| [15] |
Zhang M, Liu YH, Shi H, et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family[J]. BMC Genomics, 2018, 19(1): 159.
doi: 10.1186/s12864-018-4511-6 pmid: 29471787 |
| [16] |
Nijhawan A, Jain M, Tyagi AK, et al. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice[J]. Plant Physiol, 2008, 146(2): 333-350.
doi: 10.1104/pp.107.112821 pmid: 18065552 |
| [17] | Li HY, Li LX, ShangGuan GD, et al. Genome-wide identification and expression analysis of bZIP gene family in Carthamus tinctori-us L[J]. Sci Rep, 2020, 10(1): 15521. |
| [18] | Wang XL, Chen XL, Yang TB, et al. Genome-wide identification of bZIP family genes involved in drought and heat stresses in strawberry(Fragaria vesca)[J]. Int J Genomics, 2017, 2017: 3981031. |
| [19] | Liang Y, Xia JQ, Jiang YS, et al. Genome-wide identification and analysis of bZIP gene family and resistance of TaABI5(TabZIP96)under freezing stress in wheat(Triticum aestivum)[J]. Int J Mol Sci, 2022, 23(4): 2351. |
| [20] | Gai WX, Ma X, Qiao YM, et al. Characterization of the bZIP transcription factor family in pepper(Capsicum annuum L.): Cab-ZIP25 positively modulates the salt tolerance[J]. Front Plant Sci, 2020, 11: 139. |
| [21] |
Zhao J, Guo RR, Guo CL, et al. Evolutionary and expression analyses of the apple basic leucine zipper transcription factor family[J]. Front Plant Sci, 2016, 7: 376.
doi: 10.3389/fpls.2016.00376 pmid: 27066030 |
| [22] | Hwang I, Manoharan RK, Kang JG, et al. Genome-wide identification and characterization of bZIP transcription factors in Brassica oleracea under cold stress[J]. Biomed Res Int, 2016, 2016: 4376598. |
| [23] | Jin MJ, Gan SF, Jiao JQ, et al. Genome-wide analysis of the bZIP gene family and the role of AchnABF1 from postharvest kiwifruit(Actinidia chinensis cv. Hongyang)in osmotic and freezing stress adaptations[J]. Plant Sci, 2021, 308: 110927. |
| [24] | Zhang WH, Ye SJ, Du YL, et al. Identification and expression analysis of bZIP members under abiotic stress in mung bean(Vigna radiata)[J]. Life, 2022, 12(7): 938. |
| [25] |
Zhang Y, Gao WL, Li HT, et al. Genome-wide analysis of the bZIP gene family in Chinese jujube(Ziziphus jujuba Mill.)[J]. BMC Genomics, 2020, 21(1): 483.
doi: 10.1186/s12864-020-06890-7 pmid: 32664853 |
| [26] | Gong YY, Liu X, Chen SX, et al. Genome-wide identification and salt stress response analysis of the bZIP transcription factor family in sugar beet[J]. Int J Mol Sci, 2022, 23(19): 11573. |
| [27] | Li DY, Fu FY, Zhang HJ, et al. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato(Solanum lycopersicum L.)[J]. BMC Genomics, 2015, 16: 771. |
| [28] |
Wei KF, Chen J, Wang YM, et al. Genome-wide analysis of bZIP-encoding genes in maize[J]. DNA Res, 2012, 19(6): 463-476.
doi: 10.1093/dnares/dss026 pmid: 23103471 |
| [29] |
Lu M, Meng XX, Zhang YM, et al. Genome-wide identification and expression profiles of bZIP genes in Cannabis sativa L[J]. Cannabis Cannabinoid Res, 2022, 7(6): 882-895.
doi: 10.1089/can.2021.0153 pmid: 35020417 |
| [30] | Zhang ZR, Quan SW, Niu JX, et al. Genome-wide identification, classification, expression and duplication analysis of bZIP family genes in Juglans regia L[J]. Int J Mol Sci, 2022, 23(11): 5961. |
| [31] | Hou HY, Kong XJ, Zhou YJ, et al. Genome-wide identification and characterization of bZIP transcription factors in relation to Litchi(Litchi chinensis Sonn.) fruit ripening and postharvest storage[J]. Int J Biol Macromol, 2022, 222(Pt B): 2176-2189. |
| [32] | Pan F, Wu M, Hu WF, et al. Genome-wide identification and expression analyses of the bZIP transcription factor genes in moso bamboo(Phyllostachys edulis)[J]. Int J Mol Sci, 2019, 20(9): 2203. |
| [33] | Jin ZW, Xu W, Liu AZ. Genomic surveys and expression analysis of bZIP gene family in castor bean(Ricinus communis L.)[J]. Planta, 2014, 239(2): 299-312. |
| [34] |
Liu MY, Wen YD, Sun WJ, et al. Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartaty buckwheat[J]. BMC Genomics, 2019, 20(1): 483.
doi: 10.1186/s12864-019-5882-z pmid: 31185893 |
| [35] | Li HJ, Chen J, Zhao Q, et al. Basic leucine zipper(bZIP)transcription factor genes and their responses to drought stress in ginseng, Panax ginseng C.A. Meyer[J]. BMC Genomics, 2021, 22(1): 316. |
| [36] | Baloglu MC, Eldem V, Hajyzadeh M, et al. Genome-wide analysis of the bZIP transcription factors in cucumber[J]. PLoS One, 2014, 9(4): e96014. |
| [37] | Liu JY, Chen NN, Chen F, et al. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine(Vitis vinifera)[J]. BMC Genomics, 2014, 15: 281. |
| [38] | Wang YY, Zhang YJ, Zhou R, et al. Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame[J]. PLoS One, 2018, 13(7): e0200850. |
| [39] | Manzoor MA, Manzoor MM, Li GH, et al. Genome-wide identification and characterization of bZIP transcription factors and their expression profile under abiotic stresses in Chinese pear(Pyrus bretschneideri)[J]. BMC Plant Biol, 2021, 21(1): 413. |
| [40] | Sun MY, Fu XL, Tan QP, et al. Analysis of basic leucine zipper genes and their expression during bud dormancy in peach(Prunus persica)[J]. Plant Physiol Biochem, 2016, 104: 54-70. |
| [41] | Rong SY, Wu ZY, Cheng ZZ, et al. Genome-wide identification, evolutionary patterns, and expression analysis of bZIP gene family in olive(Olea europaea L.)[J]. Genes, 2020, 11(5): 510. |
| [42] | Zhang BY, Feng C, Chen L, et al. Identification and functional analysis of bZIP genes in cotton response to drought stress[J]. Int J Mol Sci, 2022, 23(23): 14894. |
| [43] | Jiang M, Wang Z, Ren WC, et al. Identification of the bZIP gene family and regulation of metabolites under salt stress in isatis indi-gotica[J]. Front Plant Sci, 2022, 13: 1011616. |
| [44] | Wang P, Li YX, Zhang TT, et al. Identification of the bZIP gene family and investigation of their response to drought stress in Den-drobium catenatum[J]. Agronomy, 2023, 13(1): 236. |
| [45] | Liu SY, Zhang CB, Zhu QW, et al. Genome- and transcriptome-wide systematic characterization of bZIP transcription factor family identifies promising members involved in abiotic stress response in sweetpotato[J]. Sci Hortic, 2022, 303: 111185. |
| [46] | Xi DX, Yin T, Han PC, et al. Genome-wide identification of sweet orange WRKY transcription factors and analysis of their expression in response to infection by Penicillium digitatum[J]. Curr Issues Mol Biol, 2023, 45(2): 1250-1271. |
| [47] |
Dröge-Laser W, Snoek BL, Snel B, et al. The Arabidopsis bZIP transcription factor family-an update[J]. Curr Opin Plant Biol, 2018, 45(Pt A): 36-49.
doi: S1369-5266(17)30215-7 pmid: 29860175 |
| [48] |
Mittal A, Gampala SSL, Ritchie GL, et al. Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation[J]. Plant Biotechnol J, 2014, 12(5): 578-589.
doi: 10.1111/pbi.12162 pmid: 24483851 |
| [49] | Jiang H, Gu SY, Li K, et al. Two TGA transcription factor members from hyper-ausceptible aoybean wxhibiting aignificant basal resistance to Soybean mosaic virus[J]. Int J Mol Sci, 2021, 22(21): 11329. |
| [50] |
Yu Y, Qian YC, Jiang MY, et al. Regulation mechanisms of plant basic leucine zippers to various abiotic stresses[J]. Front Plant Sci, 2020, 11: 1258.
doi: 10.3389/fpls.2020.01258 pmid: 32973828 |
| [51] |
Meng D, Cao HY, Yang Q, et al. SnRK1 kinase-mediated phosphorylation of transcription factor bZIP39 regulates sorbitol metabolism in apple[J]. Plant Physiol, 2023, 192(3): 2123-2142.
doi: 10.1093/plphys/kiad226 pmid: 37067900 |
| [52] | Muñiz García MN, Giammaria V, Grandellis C, et al. Characterization of StABF1, a stress-responsive bZIP transcription factor from Solanum tuberosum L. that is phosphorylated by StCDPK2 in vitro[J]. Planta, 2012, 235(4): 761-778. |
| [53] | An JP, Yao JF, Xu RR, et al. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation[J]. Plant Cell Environ, 2018, 41(11): 2678-2692. |
| [54] |
Zong W, Tang N, Yang J, et al. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes[J]. Plant Physiol, 2016, 171(4): 2810-2825.
doi: 10.1104/pp.16.00469 pmid: 27325665 |
| [55] | An JP, Zhang XW, Liu YJ, et al. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple[J]. J Exp Bot, 2021, 72(4): 1460-1472. |
| [56] | Han XY, Mao LC, Lu WJ, et al. Positive regulation of the transcription of AchnKCS by a bZIP transcription factor in response to ABA-stimulated suberization of kiwifruit[J]. J Agric Food Chem, 2019, 67(26): 7390-7398. |
| [57] | Zhang LN, Xie JQ, Wang LT, et al. Wheat TabZIP8, 9, 13 participate in ABA biosynthesis in NaCl-stressed roots regulated by TaCDPK9-1[J]. Plant Physiol Biochem, 2020, 151: 650-658. |
| [58] |
Gupta A, Rico-Medina A, Caño-Delgado AI. The physiology of plant responses to drought[J]. Science, 2020, 368(6488): 266-269.
doi: 10.1126/science.aaz7614 pmid: 32299946 |
| [59] | Lan YG, Pan F, Zhang KM, et al. PhebZIP47, a bZIP transcription factor from moso bamboo(Phyllostachys edulis), positively regulates the drought tolerance of transgenic plants[J]. Ind Crops Prod, 2023, 197: 116538. |
| [60] | Niu SK, Gu XY, Zhang Q, et al. Grapevine bZIP transcription factor bZIP45 regulates VvANN1 and confers drought tolerance in Arabidopsis[J]. Front Plant Sci, 2023, 14: 1128002. |
| [61] | Xiangpo Qiu WW, Yang YX, Kim HS, et al. Isolation, expression and function analysis of a bZIP transcription factor IbbZIP37 in sweetpotato(Ipomoea batatas L.[Lam])[J]. Emir J Food Agric, 2019: 134. |
| [62] | Liu CT, Mao BG, Ou SJ, et al. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice[J]. Plant Mol Biol, 2014, 84(1/2): 19-36. |
| [63] |
Yang SQ, Xu K, Chen SJ, et al. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice[J]. BMC Plant Biol, 2019, 19(1): 260.
doi: 10.1186/s12870-019-1872-1 pmid: 31208338 |
| [64] | Chen H, Chen W, Zhou JL, et al. Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice[J]. Plant Sci, 2012, 193/194: 8-17. |
| [65] |
Liu CT, Wu YB, Wang XP. bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice[J]. Planta, 2012, 235(6): 1157-1169.
doi: 10.1007/s00425-011-1564-z pmid: 22189955 |
| [66] | Pan YL, Hu X, Li CY, et al. SlbZIP38, a tomato bZIP family gene downregulated by abscisic acid, is a negative regulator of drought and salt stress tolerance[J]. Genes, 2017, 8(12): 402. |
| [67] | Yang Y, Yu TF, Ma J, et al. The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants[J]. Int J Mol Sci, 2020, 21(2): 670. |
| [68] |
Joo H, Baek W, Lim CW, et al. Post-translational modifications of bZIP transcription factors in abscisic acid signaling and drought responses[J]. Curr Genomics, 2021, 22(1): 4-15.
doi: 10.2174/1389202921999201130112116 pmid: 34045920 |
| [69] | Liu HT, Tang X, Zhang N, et al. Role of bZIP transcription factors in plant salt stress[J]. Int J Mol Sci, 2023, 24(9): 7893. |
| [70] | Bi CX, Yu YH, Dong CH, et al. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat[J]. Plant Biotechnol J, 2021, 19(2): 209-211. |
| [71] |
Wang CL, Lu GQ, Hao YQ, et al. ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton[J]. Planta, 2017, 246(3): 453-469.
doi: 10.1007/s00425-017-2704-x pmid: 28474114 |
| [72] |
Wang ZH, Yan LY, Wan LY, et al. Genome-wide systematic characterization of bZIP transcription factors and their expression profiles during seed development and in response to salt stress in peanut[J]. BMC Genomics, 2019, 20(1): 51.
doi: 10.1186/s12864-019-5434-6 pmid: 30651065 |
| [73] | Zhao P, Ye MH, Wang RQ, et al. Systematic identification and functional analysis of potato(Solanum tuberosum L.) bZIP transcription factors and overexpression of potato bZIP transcription factor StbZIP-65 enhances salt tolerance[J]. Int J Biol Macromol, 2020, 161: 155-167. |
| [74] | Kang C, Zhai H, He SZ, et al. A novel sweetpotato bZIP transcription factor gene, IbbZIP1, is involved in salt and drought tolerance in transgenic Arabidopsis[J]. Plant Cell Rep, 2019, 38(11): 1373-1382. |
| [75] | Li Q, Zhao HX, Wang XL, et al. Tartary buckwheat transcription factor FtbZIP5, regulated by FtSnRK2.6, can improve salt/drought resistance in transgenic Arabidopsis[J]. Int J Mol Sci, 2020, 21(3): 1123. |
| [76] | Zhang M, Liu YH, Cai HY, et al. The bZIP transcription factor GmbZIP15 negatively regulates salt- and drought-stress responses in soybean[J]. Int J Mol Sci, 2020, 21(20): 7778. |
| [77] | Jahed KR, Saini AK, Sherif SM. Coping with the cold: unveiling cryoprotectants, molecular signaling pathways, and strategies for cold stress resilience[J]. Front Plant Sci, 2023, 14: 1246093. |
| [78] | Li MY, Hwarari D, Li Y, et al. The bZIP transcription factors in Liriodendron chinense: Genome-wide recognition, characteristics and cold stress response[J]. Front Plant Sci, 2022, 13: 1035627. |
| [79] | Ma M, Chen QM, Dong HZ, et al. Genome-wide identification and expression analysis of the bZIP transcription factors, and functional analysis in response to drought and cold stresses in pear(Pyrus breschneideri)[J]. BMC Plant Biol, 2021, 21(1): 583. |
| [80] | Yang YX, Li JW, Li H, et al. The bZIP gene family in watermelon: genome-wide identification and expression analysis under cold stress and root-knot nematode infection[J]. PeerJ, 2019, 7: e7878. |
| [81] | Bai HR, Liao XQ, Li X, et al. DgbZIP3 interacts with DgbZIP2 to increase the expression of DgPOD for cold stress tolerance in chrysanthemum[J]. Hortic Res, 2022, 9: uhac105. |
| [82] | Li ZY, Fu DY, Wang X, et al. The transcription factor bZIP68 negatively regulates cold tolerance in maize[J]. Plant Cell, 2022, 34(8): 2833-2851. |
| [83] | Wang L, Cao HL, Qian WJ, et al. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic Arabidopsis[J]. Ann Bot, 2017, 119(7): 1195-1209. |
| [84] | Khan AH, Min L, Ma YZ, et al. High-temperature stress in crops: male sterility, yield loss and potential remedy approaches[J]. Plant Biotechnol J, 2023, 21(4): 680-697. |
| [85] | Cui Y, Ouyang SN, Zhao YJ, et al. Plant responses to high temperature and drought: a bibliometrics analysis[J]. Front Plant Sci, 2022, 13: 1052660. |
| [86] | Zheng W, Xie TT, Yu XF, et al. Characterization of bZIP transcription factors from Dimocarpus longan Lour. and analysis of their tissue-specific expression patterns and response to heat stress[J]. J Genet, 2020, 99: 69. |
| [87] |
Wang WW, Wang YF, Zhang SM, et al. Genome-wide analysis of the abiotic stress-related bZIP family in switchgrass[J]. Mol Biol Rep, 2020, 47(6): 4439-4454.
doi: 10.1007/s11033-020-05561-w pmid: 32476099 |
| [88] | Wiese AJ, Steinbachová L, Timofejeva L, et al. Arabidopsis bZIP18 and bZIP52 accumulate in nuclei following heat stress where they regulate the expression of a similar set of genes[J]. Int J Mol Sci, 2021, 22(2): 530. |
| [89] |
Zhang X, Wollenweber B, Jiang D, et al. Water deficits and heat shock effects on photosynthesis of a transgenic Arabidopsis thaliana constitutively expressing ABP9, a bZIP transcription factor[J]. J Exp Bot, 2008, 59(4): 839-848.
doi: 10.1093/jxb/erm364 pmid: 18272919 |
| [90] | Xiao YT, Chu L, Zhang YM, et al. HY5: a pivotal regulator of light-dependent development in higher plants[J]. Front Plant Sci, 2022, 12: 800989. |
| [91] | Bhagat PK, Verma D, Sharma D, et al. HY5 and ABI5 transcription factors physically interact to fine tune light and ABA signaling in Arabidopsis[J]. Plant Mol Biol, 2021, 107(1/2): 117-127. |
| [92] | Liu X, Yuan M, Dang SZ, et al. Comparative transcriptomic analysis of transcription factors and hormones during flower bud differentiation in ‘Red Globe’ grape under red-blue light[J]. Sci Rep, 2023, 13(1): 8932. |
| [93] |
Norén Lindbäck L, Ji Y, Cervela-Cardona L, et al. An interplay between bZIP16, bZIP68, and GBF1 regulates nuclear photosynthetic genes during photomorphogenesis in Arabidopsis[J]. New Phytol, 2023, 240(3): 1082-1096.
doi: 10.1111/nph.19219 pmid: 37602940 |
| [94] | Lu ZC, Qiu WM, Jin KM, et al. Identification and analysis of bZIP family genes in Sedum plumbizincicola and their potential roles in response to cadmium stress[J]. Front Plant Sci, 2022, 13: 859386. |
| [95] | Chai MN, Fan RB, Huang YM, et al. GmbZIP152, a soybean bZIP transcription factor, confers multiple biotic and abiotic stress responses in plant[J]. Int J Mol Sci, 2022, 23(18): 10935. |
| [96] | Hou FX, Liu K, Zhang N, et al. Association mapping uncovers maize ZmbZIP107 regulating root system architecture and lead absorption under lead stress[J]. Front Plant Sci, 2022, 13: 1015151. |
| [97] | Nawaz M, Sun JF, Shabbir S, et al. A review of plants strategies to resist biotic and abiotic environmental stressors[J]. Sci Total Environ, 2023, 900: 165832. |
| [98] | Baillo EH, Kimotho RN, Zhang ZB, et al. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement[J]. Genes, 2019, 10(10): 771. |
| [99] | Yang SS, Zhang XX, Zhang XM, et al. A bZIP transcription factor, PqbZIP1, is involved in the plant defense response of American ginseng[J]. PeerJ, 2022, 10: e12939. |
| [100] |
Alves MS, Dadalto SP, Gonçalves AB, et al. Plant bZIP transcription factors responsive to pathogens: a review[J]. Int J Mol Sci, 2013, 14(4): 7815-7828.
doi: 10.3390/ijms14047815 pmid: 23574941 |
| [101] | Chacón-Cerdas R, Barboza-Barquero L, Albertazzi FJ, et al. Transcription factors controlling biotic stress response in potato plants[J]. Physiol Mol Plant Pathol, 2020, 112: 101527. |
| [102] | Zhang M, Liu YH, Li ZX, et al. The bZIP transcription factor GmbZIP15 facilitates resistance against Sclerotinia sclerotiorum and Phytophthora sojae infection in soybean[J]. iScience, 2021, 24(6): 102642. |
| [103] | Lim CW, Baek W, Lim S, et al. Expression and functional roles of the pepper pathogen-induced bZIP transcription factor CabZIP2 in enhanced disease resistance to bacterial pathogen infection[J]. Mol Plant Microbe Interact, 2015, 28(7): 825-833. |
| [104] | Li DD, Li XM, Liu XT, et al. Comprehensive analysis of bZIP gene family and function of RcbZIP17 on Botrytis resistance in rose(Rosa chinensis)[J]. Gene, 2023, 849: 146867. |
| [105] | Li XL, Fan SH, Hu W, et al. Two cassava basic leucine zipper(bZIP)transcription factors(MebZIP3 and MebZIP5)confer disease resistance against cassava bacterial blight[J]. Front Plant Sci, 2017, 8: 2110. |
| [106] | An PP, Li X, Liu TX, et al. The identification of broomcorn millet bZIP transcription factors, which regulate growth and development to enhance stress tolerance and seed germination[J]. Int J Mol Sci, 2022, 23(12): 6448. |
| [107] |
Lilay GH, Castro PH, Guedes JG, et al. Rice F-bZIP transcription factors regulate the zinc deficiency response[J]. J Exp Bot, 2020, 71(12): 3664-3677.
doi: 10.1093/jxb/eraa115 pmid: 32133499 |
| [108] |
Ma HZ, Liu C, Li ZX, et al. ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development[J]. Plant Physiol, 2018, 178(2): 753-770.
doi: 10.1104/pp.18.00436 pmid: 30126870 |
| [109] | Jia ZY, Zhang MJ, Ma C, et al. Identification and functional validation of auxin-responsive Tabzip genes from wheat leaves in Arabidopsis[J]. Int J Mol Sci, 2023, 24(1): 756. |
| [110] | Martignago D, da Silveira Falavigna V, Lombardi A, et al. The bZIP transcription factor AREB3 mediates FT signalling and floral transition at the Arabidopsis shoot apical meristem[J]. PLoS Genet, 2023, 19(5): e1010766. |
| [111] | Song S, Wang GF, Wu H, et al. OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice[J]. Plant J, 2020, 103(2): 532-546. |
| [112] | Utsugi S, Ashikawa I, Nakamura S, et al. TaABI5, a wheat homolog of Arabidopsis thaliana ABA insensitive 5, controls seed germination[J]. J Plant Res, 2020, 133(2): 245-256. |
| [113] |
Han H, Wang CN, Yang XY, et al. Role of bZIP transcription factors in the regulation of plant secondary metabolism[J]. Planta, 2023, 258(1): 13.
doi: 10.1007/s00425-023-04174-4 pmid: 37300575 |
| [114] | Zhou P, Li JW, et al. Analysis of bZIP gene family in lotus(Nelumbo)and functional study of NnbZIP36 in regulating anthocyanin synthesis[J]. BMC Plant Biol, 2023, 23(1): 429. |
| [115] |
杨艳, 胡洋, 刘霓如, 等. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2):146-159.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0676 |
| Yang Y, Hu Y, Liu NR, et al. Cloning and functional analysis of MbbZIP43 gene in ‘Hongmantang’ Red-flesh apple[J]. Biotechnol bulletin, 2024, 40(2):146-159. | |
| [116] | Wang H, Xu KX, Li XG, et al. A pear S1-bZIP transcription factor PpbZIP44 modulates carbohydrate metabolism, amino acid, and flavonoid accumulation in fruits[J]. Hortic Res, 2023, 10(8): uhad140. |
| [117] | Liu DF, Shi SP, Hao ZJ, et al. OsbZIP81, A homologue of Arabidopsis VIP1, may positively regulate JA levels by directly targetting the genes in JA signaling and metabolism pathway in rice[J]. Int J Mol Sci, 2019, 20(9): 2360. |
| [118] | 吕薇, 徐郅卓, 何杰芳, 等. bZIP转录因子影响植物次生代谢研究进展[J]. 分子植物育种, 2022:1-13. |
| Lv W, Xu ZZ, He JF, et al. Research advance and role of bZIP in the biosynthesis of plant secondary metabolites[J]. Molecular Plant Breeding, 2022:1-13. |
| [1] | ZHAO Ping-juan, LIN Chen-yu, WANG Meng-yue, ZHANG Xiu-chun, LI Shu-xia, RUAN Meng-bin. Sequence Analysis of MeSDH Protein and Its Relationship with MeH1.2 in Cassava [J]. Biotechnology Bulletin, 2024, 40(9): 74-81. |
| [2] | WU Juan, WU Xiao-juan, WANG Pei-jie, XIE Rui, NIE Hu-shuai, LI Nan, MA Yan-hong. Screening and Expression Analysis of ERF Gene Related to Anthocyanin Synthesis in Colored Potato [J]. Biotechnology Bulletin, 2024, 40(9): 82-91. |
| [3] | LI Yu-qing, WU Nan, LUO Jian-rang. Cloning and Functional Analysis of bHLH Gene Related to Anthocyanin Synthesis in Paeonia qiui [J]. Biotechnology Bulletin, 2024, 40(8): 174-185. |
| [4] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [5] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| [6] | ZHANG Di, JU Rui, LI Li-mei, WANG Yu-qian, CHEN Rui, WANG Xin-yi. Application of Transcription Factor-based Biosensors in Environmental Analysis [J]. Biotechnology Bulletin, 2024, 40(6): 114-125. |
| [7] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [8] | WANG Qiu-yue, DUAN Peng-liang, LI Hai-xiao, LIU Ning, CAO Zhi-yan, DONG Jin-gao. Construction of cDNA Library of Setosphaeria turcica and Screening of Transcription Factor StMR1 Interacting Proteins [J]. Biotechnology Bulletin, 2024, 40(6): 281-289. |
| [9] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [10] | WANG Di ZHANG Xiao-yu SONG Yu-xin ZHENG Dong-ran TIAN Jing LI Yu-hua WANG Yu WU Hao. Advances in the Molecular Mechanisms of Plant Tissue Culture and Regeneration Regulated by Totipotency-related Transcription Factors [J]. Biotechnology Bulletin, 2024, 40(6): 23-33. |
| [11] | LI Hui, WEN Yu-fang, WANG Yue, JI Chao, SHI Guo-you, LUO Ying, ZHOU Yong, LI Zhi-min, WU Xiao-yu, YANG You-xin, LIU Jian-ping. Expression Characteristics and Functions of CaPIF4 in Capsicum annuum Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(4): 148-158. |
| [12] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [13] | GUO Chun, SONG Gui-mei, YAN Yan, DI Peng, WANG Ying-ping. Genome Wide Identification and Expression Analysis of the bZIP Gene Family in Panax quinquefolius [J]. Biotechnology Bulletin, 2024, 40(4): 167-178. |
| [14] | HUA Zi-qing, ZHOU Jing-yuan, DONG He-zhong. Development of Hypocotyls and Apical Hooks in Dicotyledons and Their Regulatory Mechanisms for Seedling Emergence [J]. Biotechnology Bulletin, 2024, 40(4): 23-32. |
| [15] | XIE Qian, JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi. Regulatory Genes Mining Related to Transcriptome Sequencing and Phenolic Metabolism Pathway of Canarium album Fruit with Different Fresh Food Quality [J]. Biotechnology Bulletin, 2024, 40(3): 215-228. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||