








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (10): 9-18.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0669
Previous Articles Next Articles
HE Wen-chuang1( ), XU Qiang1, QIAN Qian1,2,3(
), XU Qiang1, QIAN Qian1,2,3( ), SHANG Lian-guang1,2(
), SHANG Lian-guang1,2( )
)
Received:2024-07-14
Online:2024-10-26
Published:2024-11-20
Contact:
QIAN Qian, SHANG Lian-guang
E-mail:hewenchuang@caas.cn;qianqian@caas.cn;shanglianguang@caas.cn
HE Wen-chuang, XU Qiang, QIAN Qian, SHANG Lian-guang. Development and Prospects of Rice Pan-genomics: Important Tools and Applications[J]. Biotechnology Bulletin, 2024, 40(10): 9-18.
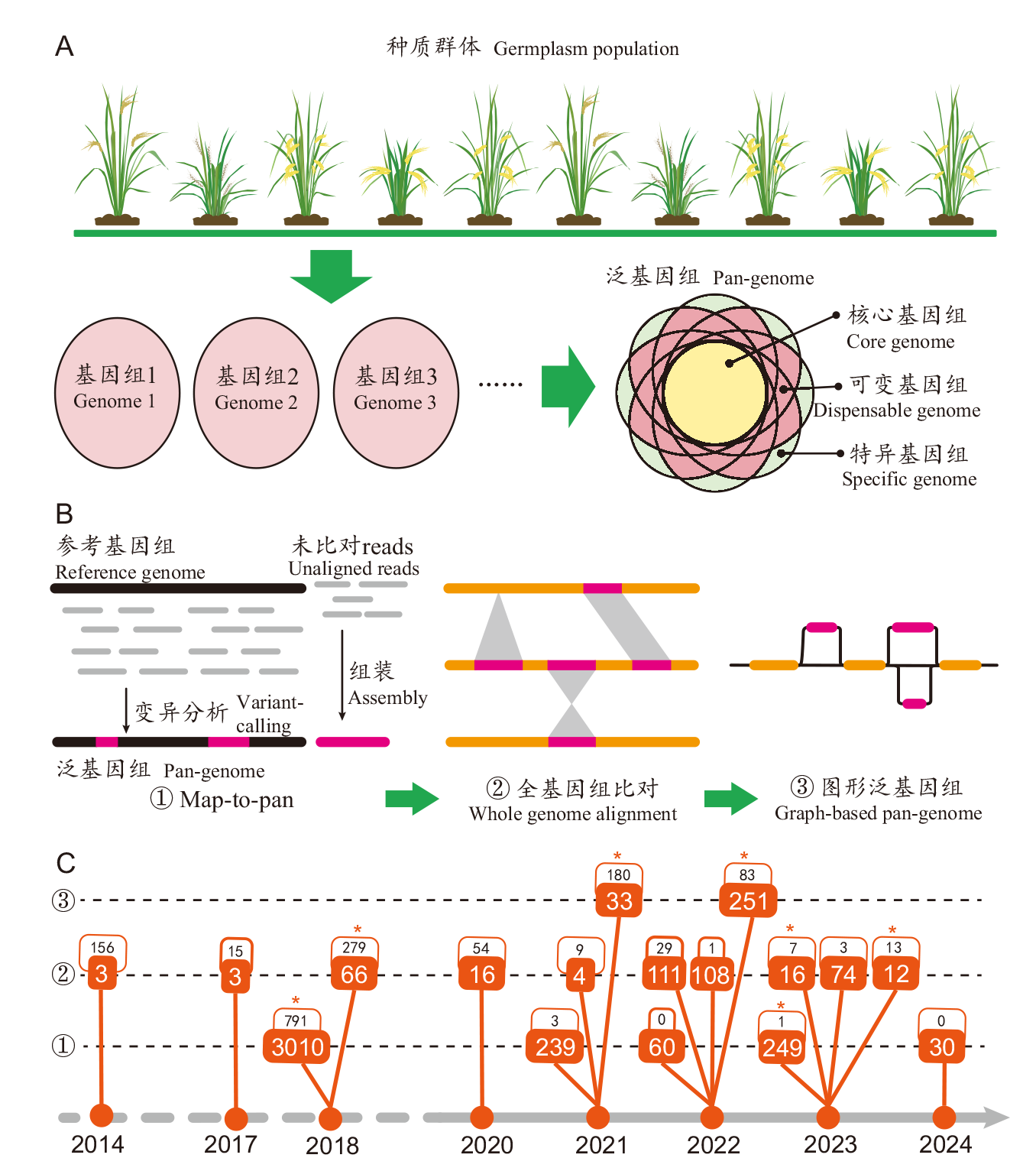
Fig. 1 Development of rice pan-genomics A: Illustration of the pan-genome definition. B: Development of pan-genome construction methods and constituent forms. C: Development timeline of rice pan-genomics studies. Each branch of the timeline iindicates a pan-genomic dataset, with two numbers showing its corresponding population size(bottom)and the number of citations to non-review papers(top). * indicates the production of a publicly accessible database or friendly analytical tool for the pan-genomic study. Number of citations are from the Web of Science database as of June 17, 2024
| 构建方法 Construction method | 发表时间 Published date | 群体大小 Population size | 群体组成 Species | 测序数据Sequencing data | 单基因组资源Genome resource | 泛基因组资源Pan-genome resource | 泛基因资源Pan-gene resource | 变异资源 Variant resource | 数据库 Database | 潜在应用 Potential application | 参考文献 Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 单基因组参考 A single genomic reference | 泛基因组参考Pan-genomic reference | GWAS等研究GWAS etc. | 单倍型分析 Haplotype analysis | |||||||||||
| Map-to-pan | 2018 | 3 010 | Os | Illumina | × | √ | √ | SNP/InDel/ SV | √ | × | √ | √ | √ | [ |
| 2021 | 239 | Osj | Illumina | × | √ | √ | SNPs/PAV | × | × | √ | √ | √ | [ | |
| 2023 | 60 | Os | Illumina | × | √ | √ | SNP/PAV | × | × | √ | × | √ | [ | |
| 2024 | 249 | Ogla/Ob | Illumina | × | √ | √ | 无 | × | × | √ | × | × | [ | |
| 2024 | 30 | Oa/ Ogra/Ol | Illumina | × | √ | √ | SNP/PAV | × | × | √ | × | √ | [ | |
| 全基因组比对Whole genome alignment | 2014 | 3 | Os | Illumina | √ | × | 冗余 | 无 | × | √ | × | × | √ | [ |
| 2017 | 3 | Ogla | Illumina | √ | × | 冗余 | 无 | × | √ | × | × | √ | [ | |
| 2018 | 66 | Os/Or | Illumina | √ | × | √ | SNP/InDel | √ | √ | × | × | √ | [ | |
| 2020 | 16 | Os | Illumina/Pacbio/Bionano | √ | × | × | InDel/SV | × | √ | × | × | √ | [ | |
| 2021 | 4 | Os | Illumina/Pacbio | √ | × | √ | SNP/InDel/SV | × | √ | × | × | √ | [ | |
| 2022 | 111 | Or/Os | Illumina/Nanopore | √ | √ | √ | SV | × | √ | √ | × | √ | [ | |
| 2022 | 108 | Os | Illumina | √ | √ | √ | InDel/PAV | × | √ | √ | × | √ | [ | |
| 2023 | 74 | Or/Os/杂草稻 | PacBio(12)/Hi-C(4) | √ | × | √ | SNP/PAV | × | √ | × | × | √ | [ | |
| 2023 | 16 | Os | Iso-Seq/RNA-Seq | [16]* | × | √ | 可变剪切事件 | √ | √ | × | × | √ | [ | |
| 2023 | 12 | Os/Ogla | [21]* | [21]* | √ | √ | PAV | × | √ | √ | × | √ | [ | |
| 图形泛基因组Graph-based pan-genome | 2021 | 33 | Os/Ogla | Illumina/Pacbio/Bionano(3) | √ | √ | √ | SV/gCNV | √ | √ | √ | × | √ | [ |
| 2022 | 251 | Os/Or/ Ogla/Ob | Illumina/Nanopore/Hi-C(4) | √ | √ | √ | SV | √ | √ | √ | √ | √ | [ | |
Table 1 Summary of published pan-genome studies in rice
| 构建方法 Construction method | 发表时间 Published date | 群体大小 Population size | 群体组成 Species | 测序数据Sequencing data | 单基因组资源Genome resource | 泛基因组资源Pan-genome resource | 泛基因资源Pan-gene resource | 变异资源 Variant resource | 数据库 Database | 潜在应用 Potential application | 参考文献 Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 单基因组参考 A single genomic reference | 泛基因组参考Pan-genomic reference | GWAS等研究GWAS etc. | 单倍型分析 Haplotype analysis | |||||||||||
| Map-to-pan | 2018 | 3 010 | Os | Illumina | × | √ | √ | SNP/InDel/ SV | √ | × | √ | √ | √ | [ |
| 2021 | 239 | Osj | Illumina | × | √ | √ | SNPs/PAV | × | × | √ | √ | √ | [ | |
| 2023 | 60 | Os | Illumina | × | √ | √ | SNP/PAV | × | × | √ | × | √ | [ | |
| 2024 | 249 | Ogla/Ob | Illumina | × | √ | √ | 无 | × | × | √ | × | × | [ | |
| 2024 | 30 | Oa/ Ogra/Ol | Illumina | × | √ | √ | SNP/PAV | × | × | √ | × | √ | [ | |
| 全基因组比对Whole genome alignment | 2014 | 3 | Os | Illumina | √ | × | 冗余 | 无 | × | √ | × | × | √ | [ |
| 2017 | 3 | Ogla | Illumina | √ | × | 冗余 | 无 | × | √ | × | × | √ | [ | |
| 2018 | 66 | Os/Or | Illumina | √ | × | √ | SNP/InDel | √ | √ | × | × | √ | [ | |
| 2020 | 16 | Os | Illumina/Pacbio/Bionano | √ | × | × | InDel/SV | × | √ | × | × | √ | [ | |
| 2021 | 4 | Os | Illumina/Pacbio | √ | × | √ | SNP/InDel/SV | × | √ | × | × | √ | [ | |
| 2022 | 111 | Or/Os | Illumina/Nanopore | √ | √ | √ | SV | × | √ | √ | × | √ | [ | |
| 2022 | 108 | Os | Illumina | √ | √ | √ | InDel/PAV | × | √ | √ | × | √ | [ | |
| 2023 | 74 | Or/Os/杂草稻 | PacBio(12)/Hi-C(4) | √ | × | √ | SNP/PAV | × | √ | × | × | √ | [ | |
| 2023 | 16 | Os | Iso-Seq/RNA-Seq | [16]* | × | √ | 可变剪切事件 | √ | √ | × | × | √ | [ | |
| 2023 | 12 | Os/Ogla | [21]* | [21]* | √ | √ | PAV | × | √ | √ | × | √ | [ | |
| 图形泛基因组Graph-based pan-genome | 2021 | 33 | Os/Ogla | Illumina/Pacbio/Bionano(3) | √ | √ | √ | SV/gCNV | √ | √ | √ | × | √ | [ |
| 2022 | 251 | Os/Or/ Ogla/Ob | Illumina/Nanopore/Hi-C(4) | √ | √ | √ | SV | √ | √ | √ | √ | √ | [ | |

Fig. 2 Distribution of major fields of non-review citations for rice pan-genome studies * statistical results of MeSH Headings Keywords in the WOS database(the total number of literatures participating in the statistics n=859, the number of citations for a single entry ≥50). Only the results of terms related to important fields of rice are shown here, and only one keyword describing similar or similar fields is shown

Fig. 3 Main applications of rice pan-genomics tools The elements in the evolution and domestication diagram are modified according to Chen et al[25]. The elements in the diagram for biological breeding and intelligent breeding are modified according to Ferrero-Serrano et al[26]
| [1] | Tettelin H, Masignani V, Cieslewicz MJ, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”[J]. Proc Natl Acad Sci U S A, 2005, 102(39): 13950-13955. |
| [2] |
Li RQ, Li YR, Zheng HC, et al. Building the sequence map of the human pan-genome[J]. Nat Biotechnol, 2010, 28(1): 57-63.
doi: 10.1038/nbt.1596 pmid: 19997067 |
| [3] |
Li YH, Zhou GY, Ma JX, et al. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits[J]. Nat Biotechnol, 2014, 32(10): 1045-1052.
doi: 10.1038/nbt.2979 |
| [4] |
Bayer PE, Golicz AA, Scheben A, et al. Plant pan-genomes are the new reference[J]. Nat Plants, 2020, 6(8): 914-920.
doi: 10.1038/s41477-020-0733-0 pmid: 32690893 |
| [5] | Shi JP, Tian ZX, Lai JS, et al. Plant pan-genomics and its applications[J]. Mol Plant, 2023, 16(1): 168-186. |
| [6] |
Zhang F, Xue HZ, Dong XR, et al. Long-read sequencing of 111 rice genomes reveals significantly larger pan-genomes[J]. Genome Res, 2022, 32(5): 853-863.
doi: 10.1101/gr.276015.121 pmid: 35396275 |
| [7] |
Wang J, Yang W, Zhang SH, et al. A pangenome analysis pipeline provides insights into functional gene identification in rice[J]. Genome Biol, 2023, 24(1): 19.
doi: 10.1186/s13059-023-02861-9 pmid: 36703158 |
| [8] | Schatz MC, Maron LG, Stein JC, et al. Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica[J]. Genome Biol, 2014, 15(11): 506. |
| [9] | Monat C, Pera B, Ndjiondjop MN, et al. De novo assemblies of three Oryza glaberrima accessions provide first insights about pan-genome of African rices[J]. Genome Biol Evol, 2017, 9(1): 1-6. |
| [10] | Wang WS, Mauleon R, Hu ZQ, et al. Genomic variation in 3, 010 diverse accessions of Asian cultivated rice[J]. Nature, 2018, 557(7703): 43-49. |
| [11] |
Liu CX, Peng P, Li WG, et al. Deciphering variation of 239 elite Japonica rice genomes for whole genome sequences-enabled breeding[J]. Genomics, 2021, 113(5): 3083-3091.
doi: 10.1016/j.ygeno.2021.07.002 pmid: 34237377 |
| [12] | Woldegiorgis ST, Wu T, Gao LH, et al. Identification of heat-tolerant genes in non-reference sequences in rice by integrating pan-genome, transcriptomics, and QTLs[J]. Genes, 2022, 13(8): 1353. |
| [13] | Christine TD, Clothilde C, Mathieu B, et al. FrangiPANe, a tool for creating a panreference using left behind reads[J]. NAR Genom Bioinform, 2023, 5(1): lqad013. |
| [14] | Shivute FN, Zhong Y, Wu JW, et al. Genome-wide and pan-genomic analysis reveals rich variants of NBS-LRR genes in a newly developed wild rice line from Oryza alta Swallen[J]. Front Plant Sci, 2024, 15: 1345708. |
| [15] |
Zhao Q, Feng Q, Lu HY, et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice[J]. Nat Genet, 2018, 50(2): 278-284.
doi: 10.1038/s41588-018-0041-z pmid: 29335547 |
| [16] |
Zhou Y, Chebotarov D, Kudrna D, et al. A platinum standard pan-genome resource that represents the population structure of Asian rice[J]. Sci Data, 2020, 7(1): 113.
doi: 10.1038/s41597-020-0438-2 pmid: 32265447 |
| [17] |
Panibe JP, Wang L, Li J, et al. Chromosomal-level genome assembly of the semi-dwarf rice Taichung Native 1, an initiator of green revolution[J]. Genomics, 2021, 113(4): 2656-2674.
doi: 10.1016/j.ygeno.2021.06.006 pmid: 34111524 |
| [18] | Singh PK, Rawal HC, Panda AK, et al. Pan-genomic, transcriptomic, and miRNA analyses to decipher genetic diversity and anthocyanin pathway genes among the traditional rice landraces[J]. Genomics, 2022, 114(5): 110436. |
| [19] |
Wu DY, Xie LJ, Sun YQ, et al. A syntelog-based pan-genome provides insights into rice domestication and de-domestication[J]. Genome Biol, 2023, 24(1): 179.
doi: 10.1186/s13059-023-03017-5 pmid: 37537691 |
| [20] | Yu ZC, Chen YM, Zhou Y, et al. Rice Gene Index: a comprehensive pan-genome database for comparative and functional genomics of Asian rice[J]. Mol Plant, 2023, 16(5): 798-801. |
| [21] |
Qin P, Lu HW, Du HL, et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations[J]. Cell, 2021, 184(13): 3542-3558.e16.
doi: 10.1016/j.cell.2021.04.046 pmid: 34051138 |
| [22] |
Shang LG, Li XX, He HY, et al. A super pan-genomic landscape of rice[J]. Cell Res, 2022, 32(10): 878-896.
doi: 10.1038/s41422-022-00685-z pmid: 35821092 |
| [23] | Kawahara Y, de la Bastide M, Hamilton JP, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data[J]. Rice(N Y), 2013, 6(1): 4. |
| [24] | Sun C, Hu ZQ, Zheng TQ, et al. RPAN: rice pan-genome browser for -3000 rice genomes[J]. Nucleic Acids Res, 2017, 45(2): 597-605. |
| [25] | Chen WK, Chen L, Zhang X, et al. Convergent selection of a WD40 protein that enhances grain yield in maize and rice[J]. Science, 2022, 375(6587): eabg7985. |
| [26] | Ferrero-Serrano Á, Chakravorty D, Kirven KJ, et al. Oryza CLIMtools: a genome-environment association resource reveals adaptive roles for heterotrimeric G proteins in the regulation of rice agronomic traits[J]. Plant Commun, 2024, 5(4): 100813. |
| [27] | Wei H, Wang XM, Zhang ZP, et al. Uncovering key salt-tolerant regulators through a combined eQTL and GWAS analysis using the super pan-genome in rice[J]. Natl Sci Rev, 2024, 11(4): nwae043. |
| [28] |
Lv Y, Liu CC, Li XX, et al. A centromere map based on super pan-genome highlights the structure and function of rice centromeres[J]. J Integr Plant Biol, 2024, 66(2): 196-207.
doi: 10.1111/jipb.13607 |
| [29] | Cui YC, Lin YR, Wei H, et al. Identification of salt tolerance-associated presence-absence variations in the OsMADS56 gene through the integration of DEGs dataset and eQTL analysis[J]. New Phytol, 2024, 243(3): 833-838. |
| [30] |
Zhou Y, Yu ZC, Chebotarov D, et al. Pan-genome inversion index reveals evolutionary insights into the subpopulation structure of Asian rice[J]. Nat Commun, 2023, 14(1): 1567.
doi: 10.1038/s41467-023-37004-y pmid: 36944612 |
| [31] |
Wang TY, He WC, Li XX, et al. A rice variation map derived from 10 548 rice accessions reveals the importance of rare variants[J]. Nucleic Acids Res, 2023, 51(20): 10924-10933.
doi: 10.1093/nar/gkad840 pmid: 37843097 |
| [32] | Zhang H, Chen W, Zhu D, et al. Population-level exploration of alternative splicing and its unique role in controlling agronomic traits of rice[J]. Plant Cell, 2024: koae181. |
| [33] | Li XX, Dai XF, He HY, et al. A pan-TE map highlights transposable elements underlying domestication and agronomic traits in Asian rice[J]. Natl Sci Rev, 2024, 11(6): nwae188. |
| [34] |
He HY, Leng Y, Cao XL, et al. The pan-tandem repeat map highlights multiallelic variants underlying gene expression and agronomic traits in rice[J]. Nat Commun, 2024, 15: 7291.
doi: 10.1038/s41467-024-51854-0 pmid: 39181885 |
| [35] | Wang J, Hu HF, Jiang XY, et al. Pangenome-wide association study and transcriptome analysis reveal a novel QTL and candidate genes controlling both panicle and leaf blast resistance in rice[J]. Rice(N Y), 2024, 17(1): 27. |
| [36] | Singh G, Singh N, Ellur RK, et al. Genetic enhancement for biotic stress resistance in basmati rice through marker-assisted backcross breeding[J]. Int J Mol Sci, 2023, 24(22): 16081. |
| [37] | Sun Y, Zhang PT, Kou DR, et al. Terpene synthases in rice pan-genome and their responses to Chilo suppressalis larvae infesting[J]. Front Plant Sci, 2022, 13: 905982. |
| [38] |
Lin YR, Zhu YW, Cui YC, et al. Identification of natural allelic variation in TTL1 controlling thermotolerance and grain size by a rice super pan-genome[J]. J Integr Plant Biol, 2023, 65(12): 2541-2551.
doi: 10.1111/jipb.13568 |
| [39] |
Li WH, Wang DX, Hong XK, et al. Identification and validation of new MADS-box homologous genes in 3010 rice pan-genome[J]. Plant Cell Rep, 2023, 42(6): 975-988.
doi: 10.1007/s00299-023-03006-9 pmid: 37016094 |
| [40] | Daware A, Malik A, Srivastava R, et al. Rice Pangenome Genotyping Array: an efficient genotyping solution for pangenome-based accelerated genetic improvement in rice[J]. Plant J, 2023, 113(1): 26-46. |
| [41] | He WC, He HY, Yuan QL, et al. Widespread inversions shape the genetic and phenotypic diversity in rice[J]. Sci Bull, 2024, 69(5): 593-596. |
| [42] | Ramsbottom KA, Prakash A, Perez-Riverol Y, et al. Meta-analysis of rice phosphoproteomics data to understand variation in cell signaling across the rice pan-genome[J]. J Proteome Res, 2024, 23(7): 2518-2531. |
| [43] | Brotman Y, Llorente-Wiegand C, Oyong G, et al. The genetics underlying metabolic signatures in a brown rice diversity panel and their vital role in human nutrition[J]. Plant J, 2021, 106(2): 507-525. |
| [44] |
Wei X, Qiu J, Yong KC, et al. A quantitative genomics map of rice provides genetic insights and guides breeding[J]. Nat Genet, 2021, 53(2): 243-253.
doi: 10.1038/s41588-020-00769-9 pmid: 33526925 |
| [45] | Wang S, Qian YQ, Zhao RP, et al. Graph-based pan-genomes: increased opportunities in plant genomics[J]. J Exp Bot, 2023, 74(1): 24-39. |
| [46] | Chen RZ, Deng YW, Ding YL, et al. Rice functional genomics: decades’ efforts and roads ahead[J]. Sci China Life Sci, 2022, 65(1): 33-92. |
| [47] | Li HH, Li X, Zhang P, et al. Smart Breeding Platform: a web-based tool for high-throughput population genetics, phenomics, and genomic selection[J]. Mol Plant, 2024, 17(5): 677-681. |
| [1] | XIE Wei, LIU Chun-ming. Commercialization of Biological Breeding in China: Opportunities and Policy Issues [J]. Biotechnology Bulletin, 2023, 39(1): 16-20. |
| [2] | CHANG Li, DENG Xin, TANG Hui-juan, LI Jian-jun, HUANG Si-qi, CHEN An-guo, ZHANG Cui-ping, ZHAO Li-ning, LI De-fang. Research Trend and Technical Composition of Cannabis sativa Based on Patent Analysis [J]. Biotechnology Bulletin, 2018, 34(12): 195-201. |
| [3] | YANG Jing, DAI Qiu-zhong, HOU Zhen-ping, WANG Yan-zhou, WANG Man-sheng. Development Trends and Technical Hotspots in Feed Processing Technology of Bast Fiber Crops Based on the Analysis of Patents [J]. Biotechnology Bulletin, 2018, 34(12): 207-214. |
| [4] | SONG Na-na, CHAI Zhi-xin, ZHONG Jin-cheng. Research Advances on Long Non-coding RNA [J]. Biotechnology Bulletin, 2016, 32(9): 23-31. |
| [5] | Gong Jing, Liu Chunjie, Miao Xiaoping, Guo Anyuan. Research Progress of the Human Long Non-coding RNA Related SNP Identification and Function Prediction [J]. Biotechnology Bulletin, 2015, 31(11): 27-34. |
| [6] | Jin Jinpu, Guo Anyuan, He Kun, Zhang He, Zhu Qihui, Chen Xin, Gao Ge, Luo Jingchu. Classification, Prediction and Database Construction of Plant Transcription Factors [J]. Biotechnology Bulletin, 2015, 31(11): 68-77. |
| [7] | Wang Huili, Guo Anyuan. An Introduction to Metagenome Databases of Environmental Microbiology [J]. Biotechnology Bulletin, 2015, 31(11): 78-88. |
| [8] | Yang Jian, Cai Haoyang. The Cancer-related Bioinformatics Databases [J]. Biotechnology Bulletin, 2015, 31(11): 89-101. |
| [9] | Luo Jingchu. Teaching Examples of Applied Bioinformatics Course [J]. Biotechnology Bulletin, 2015, 31(11): 102-111. |
| [10] | Hao Xinning, Sun Wei, Zhang Xuefu. An Analysis of Knowledge Structure Evolution on Biological Breeding Field [J]. Biotechnology Bulletin, 2013, 29(11): 186-192. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||