








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (1): 143-156.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0576
Previous Articles Next Articles
LI Yu-xin1( ), LI Miao1,2, DU Xiao-fen1,2, HAN Kang-ni1, LIAN Shi-chao1, WANG Jun1,2(
), LI Miao1,2, DU Xiao-fen1,2, HAN Kang-ni1, LIAN Shi-chao1, WANG Jun1,2( )
)
Received:2024-06-15
Online:2025-01-26
Published:2025-01-22
Contact:
WANG Jun
E-mail:ecnulyx@163.com;128wan@163.com
LI Yu-xin, LI Miao, DU Xiao-fen, HAN Kang-ni, LIAN Shi-chao, WANG Jun. Identification and Expression Analysis of SiSAP Gene Family in Foxtail Millet(Setaria italica)[J]. Biotechnology Bulletin, 2025, 41(1): 143-156.
| 基因名称Gene name | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| SiSAP1 | CTGCTGCTGTAATCCCCAAG | AGTGGCAGTCATGTTTGTCG |
| SiSAP2 | GTTCCTCGCAATTTTCTGGA | CCAAGTACGCGTTTGCATAG |
| SiSAP3 | ACGGTATATCTTGCGCTCGT | GCACTGTTGCCGAGATGTAA |
| SiSAP4 | GCAGATAGCCAAGCAGAACC | CCACCACCTGAAGGAACCTA |
| SiSAP6 | GTCTCCTCTCGCTCATCACG | CCTCACGCTTCTTCTTCTGC |
| SiSAP7 | AAGATTCCATCCAGCAGAGC | CATCACCGCCATCATTACAC |
| SiSAP8 | ATCCGGTGTTGCTAGTGGAG | ATGGGCAAATTTGAAGTCCA |
| SiSAP11 | ATACCGGACCCAAACAACTG | ACCGTCTCAAGACAGGTTCG |
| SiSAP14 | AACCCATTTCCTCTCCCTCA | GGCTCCTCCACCTTCTTGTC |
| SiACTIN | TGATCTCACTGACAGTCTGATG | GATGTCTCTTACAATTTCCCGC |
Table 1 Primers for RT-qPCR in this study
| 基因名称Gene name | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| SiSAP1 | CTGCTGCTGTAATCCCCAAG | AGTGGCAGTCATGTTTGTCG |
| SiSAP2 | GTTCCTCGCAATTTTCTGGA | CCAAGTACGCGTTTGCATAG |
| SiSAP3 | ACGGTATATCTTGCGCTCGT | GCACTGTTGCCGAGATGTAA |
| SiSAP4 | GCAGATAGCCAAGCAGAACC | CCACCACCTGAAGGAACCTA |
| SiSAP6 | GTCTCCTCTCGCTCATCACG | CCTCACGCTTCTTCTTCTGC |
| SiSAP7 | AAGATTCCATCCAGCAGAGC | CATCACCGCCATCATTACAC |
| SiSAP8 | ATCCGGTGTTGCTAGTGGAG | ATGGGCAAATTTGAAGTCCA |
| SiSAP11 | ATACCGGACCCAAACAACTG | ACCGTCTCAAGACAGGTTCG |
| SiSAP14 | AACCCATTTCCTCTCCCTCA | GGCTCCTCCACCTTCTTGTC |
| SiACTIN | TGATCTCACTGACAGTCTGATG | GATGTCTCTTACAATTTCCCGC |
| 基因Gene | 基因号Gene ID | 基因位置Gene location | 蛋白长度Protein length/aa | 分子量Molecular weight/kD | 等电点pI | 亲水性GRAVY |
|---|---|---|---|---|---|---|
| SiSAP1 | Seita.1G049700 | Chr.1:4 826 713-4828307 | 172 | 18.38 | 7.99 | -0.216 |
| SiSAP2 | Seita.1G050100 | Chr.1:4 844 574-4846624 | 172 | 18.21 | 8.28 | -0.198 |
| SiSAP3 | Seita.1G179600 | Chr.1:25 817 455-25 819 730 | 150 | 15.65 | 9.28 | -0.289 |
| SiSAP4 | Seita.2G045100 | Chr.2:3 644 125-3 646 735 | 165 | 18.09 | 8.02 | -0.479 |
| SiSAP5 | Seita.2G188600 | Chr.2:28 469 651-28 470 203 | 184 | 19.74 | 8.75 | -0.526 |
| SiSAP6 | Seita.2G195200 | Chr.2:29 175 966-29 176 521 | 185 | 19.76 | 9.04 | -0.546 |
| SiSAP7 | Seita.2G252300 | Chr.2:35 515 181-35 516 761 | 170 | 17.55 | 8.90 | -0.234 |
| SiSAP8 | Seita.2G358600 | Chr.2:43 671 478-43 674 297 | 291 | 32.12 | 8.66 | -0.627 |
| SiSAP9 | Seita.5G232000 | Chr.5:29 446 450-29 446 969 | 173 | 17.47 | 8.96 | -0.097 |
| SiSAP10 | Seita.5G232100 | Chr.5:29 462 246-29 462 780 | 178 | 17.86 | 9.06 | -0.063 |
| SiSAP11 | Seita.5G297700 | Chr.5:35 289 769-35 291 603 | 154 | 15.86 | 8.74 | -0.352 |
| SiSAP12 | Seita.5G330400 | Chr.5:37 812 781-37 813 282 | 167 | 17.11 | 8.76 | -0.089 |
| SiSAP13 | Seita.6G159700 | Chr.6:28 237 350-28 238 434 | 256 | 26.19 | 8.47 | -0.190 |
| SiSAP14 | Seita.6G203700 | Chr.6:32 355 017-32 361 096 | 179 | 18.82 | 9.14 | -0.357 |
| SiSAP15 | Seita.7G243600 | Chr.7:30 408 518-30 409 232 | 236 | 24.02 | 8.60 | -0.373 |
| SiSAP16 | Seita.9G061100 | Chr.9:3 504 533-3 504 971 | 146 | 15.05 | 9.36 | -0.312 |
| SiSAP17 | Seita.9G316800 | Chr.9:36 489 146-36 489 611 | 155 | 16.52 | 6.62 | -0.408 |
Table 2 Identification of SiSAP gene family members
| 基因Gene | 基因号Gene ID | 基因位置Gene location | 蛋白长度Protein length/aa | 分子量Molecular weight/kD | 等电点pI | 亲水性GRAVY |
|---|---|---|---|---|---|---|
| SiSAP1 | Seita.1G049700 | Chr.1:4 826 713-4828307 | 172 | 18.38 | 7.99 | -0.216 |
| SiSAP2 | Seita.1G050100 | Chr.1:4 844 574-4846624 | 172 | 18.21 | 8.28 | -0.198 |
| SiSAP3 | Seita.1G179600 | Chr.1:25 817 455-25 819 730 | 150 | 15.65 | 9.28 | -0.289 |
| SiSAP4 | Seita.2G045100 | Chr.2:3 644 125-3 646 735 | 165 | 18.09 | 8.02 | -0.479 |
| SiSAP5 | Seita.2G188600 | Chr.2:28 469 651-28 470 203 | 184 | 19.74 | 8.75 | -0.526 |
| SiSAP6 | Seita.2G195200 | Chr.2:29 175 966-29 176 521 | 185 | 19.76 | 9.04 | -0.546 |
| SiSAP7 | Seita.2G252300 | Chr.2:35 515 181-35 516 761 | 170 | 17.55 | 8.90 | -0.234 |
| SiSAP8 | Seita.2G358600 | Chr.2:43 671 478-43 674 297 | 291 | 32.12 | 8.66 | -0.627 |
| SiSAP9 | Seita.5G232000 | Chr.5:29 446 450-29 446 969 | 173 | 17.47 | 8.96 | -0.097 |
| SiSAP10 | Seita.5G232100 | Chr.5:29 462 246-29 462 780 | 178 | 17.86 | 9.06 | -0.063 |
| SiSAP11 | Seita.5G297700 | Chr.5:35 289 769-35 291 603 | 154 | 15.86 | 8.74 | -0.352 |
| SiSAP12 | Seita.5G330400 | Chr.5:37 812 781-37 813 282 | 167 | 17.11 | 8.76 | -0.089 |
| SiSAP13 | Seita.6G159700 | Chr.6:28 237 350-28 238 434 | 256 | 26.19 | 8.47 | -0.190 |
| SiSAP14 | Seita.6G203700 | Chr.6:32 355 017-32 361 096 | 179 | 18.82 | 9.14 | -0.357 |
| SiSAP15 | Seita.7G243600 | Chr.7:30 408 518-30 409 232 | 236 | 24.02 | 8.60 | -0.373 |
| SiSAP16 | Seita.9G061100 | Chr.9:3 504 533-3 504 971 | 146 | 15.05 | 9.36 | -0.312 |
| SiSAP17 | Seita.9G316800 | Chr.9:36 489 146-36 489 611 | 155 | 16.52 | 6.62 | -0.408 |
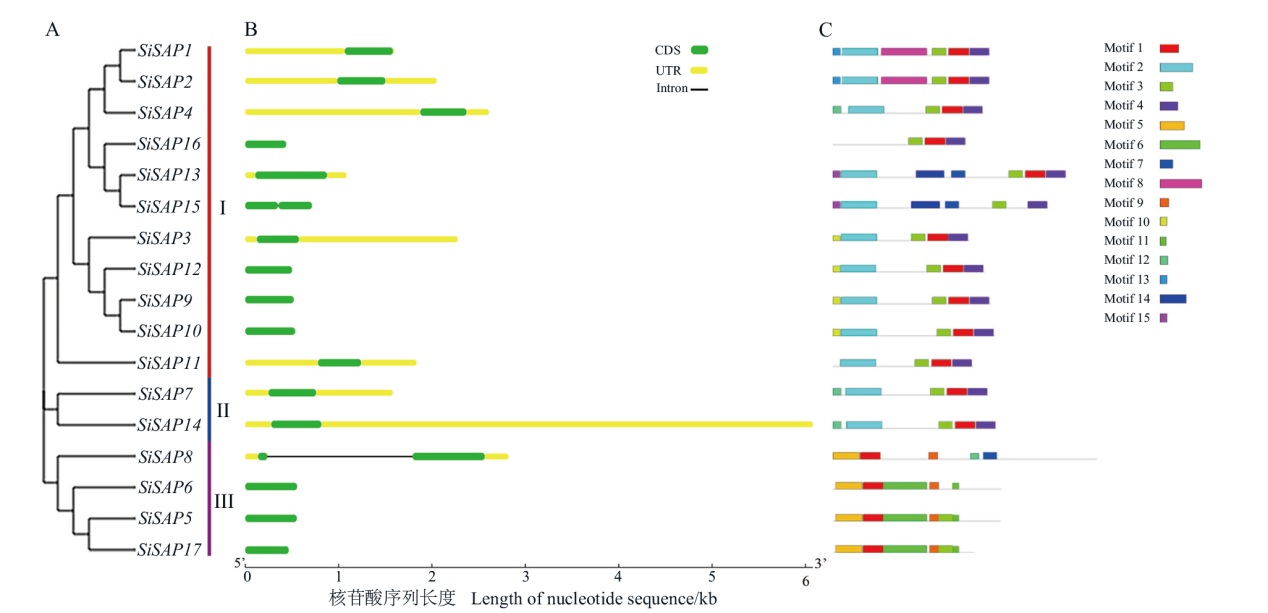
Fig. 2 Analysis of SiSAP gene family evolutionary tree, gene structure and motifs A: Phylogenetic relationship. B: Gene structure. C: Conserved motifs. Motif2 and motif14-motif7 are A20; motif3-motif1, motif5-motif1 and motif9-motif3-motif11 are AN1
| 同源基因 Homologous gene | 非同义替换率(Ka) Non-synonymous substitution | 同义替换率(Ks) Synonymous substitution | 非同义替换率/同义替换率 Ka/Ks |
|---|---|---|---|
| SiSAP4-SiSAP16 | 0.41 | 1.21 | 0.34 |
| SiSAP7-SiSAP14 | 0.20 | 0.47 | 0.42 |
Table 3 Ka/Ks ratios of segmental duplicated gene pairs in the SiSAP gene family
| 同源基因 Homologous gene | 非同义替换率(Ka) Non-synonymous substitution | 同义替换率(Ks) Synonymous substitution | 非同义替换率/同义替换率 Ka/Ks |
|---|---|---|---|
| SiSAP4-SiSAP16 | 0.41 | 1.21 | 0.34 |
| SiSAP7-SiSAP14 | 0.20 | 0.47 | 0.42 |

Fig. 6 Phylogenetic tree of SAP gene family in plants Red stars: Setaria italica. Orange checks: Oraza sativa. Blue triangles: Triticum aestivum. Green circles: Hordeum vulgare. Yellow rects: Arabidopsis thaliana. Purple squares: Tamarix hispida

Fig. 7 Prediction of cis-acting element and transcription factor binding sites in the SiSAP gene family of foxtail millet A: Analysis of cis-acting element in promoter sequence of SiSAP family genes. The different intensity colors and numbers of the grid indicate the cis-acting element numbers in the SiSAP genes. B: The different colored histogram represents the sum of the cis-acting elements in each category. C: Only the top 15 TFs that bind to the promoter of SiSAP genes are shown here. The different intensity colors and numbers of the grid indicate the numbers of putative TF sites
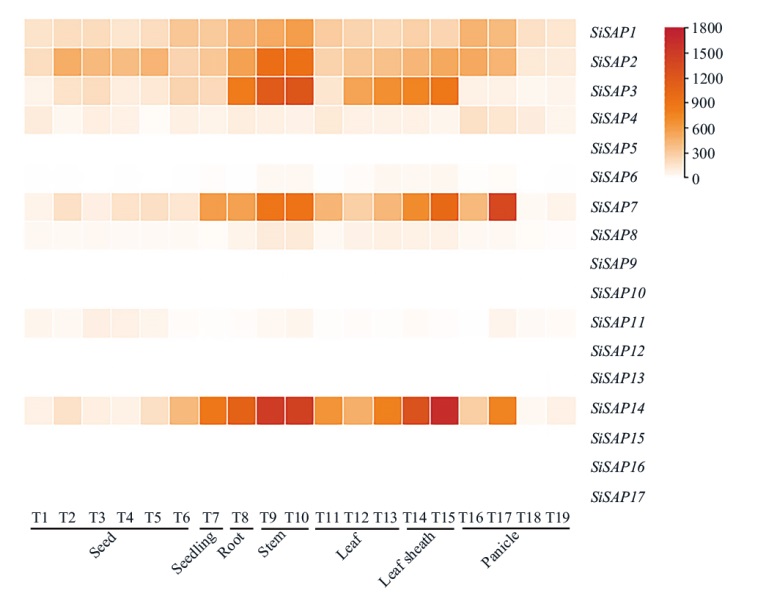
Fig. 8 Tissue expression pattern analysis of SiSAP gene family in foxtail millet T1: Immature seed at S1. T2: Immature seed at S2. T3: Immature seed at S3. T4: Immature seed at S4. T5: Immature seed at S5. T6: Seeds germinated for 3 d. T7: Plant of one-tip-two-leaf. T8: Root at filling stage. T9: Neck-panicle-internodes at filling stage. T10: Stem-top-second at filling stage. T11: Leaves after 2 d of heading(Leaf-top-2-3). T12: Flag leaf at filling stage. T13: Leaf-top-fourth at filling stage. T14: Flag leaf sheath at filling stage. T15: Leaf-sheath-top-fourth at filling stage. T16: Primary panicle at differentiation stage. T17: Third panicle at differentiation stage. T18: Immature spikelet at S2. T19: Immature spikelet at S4
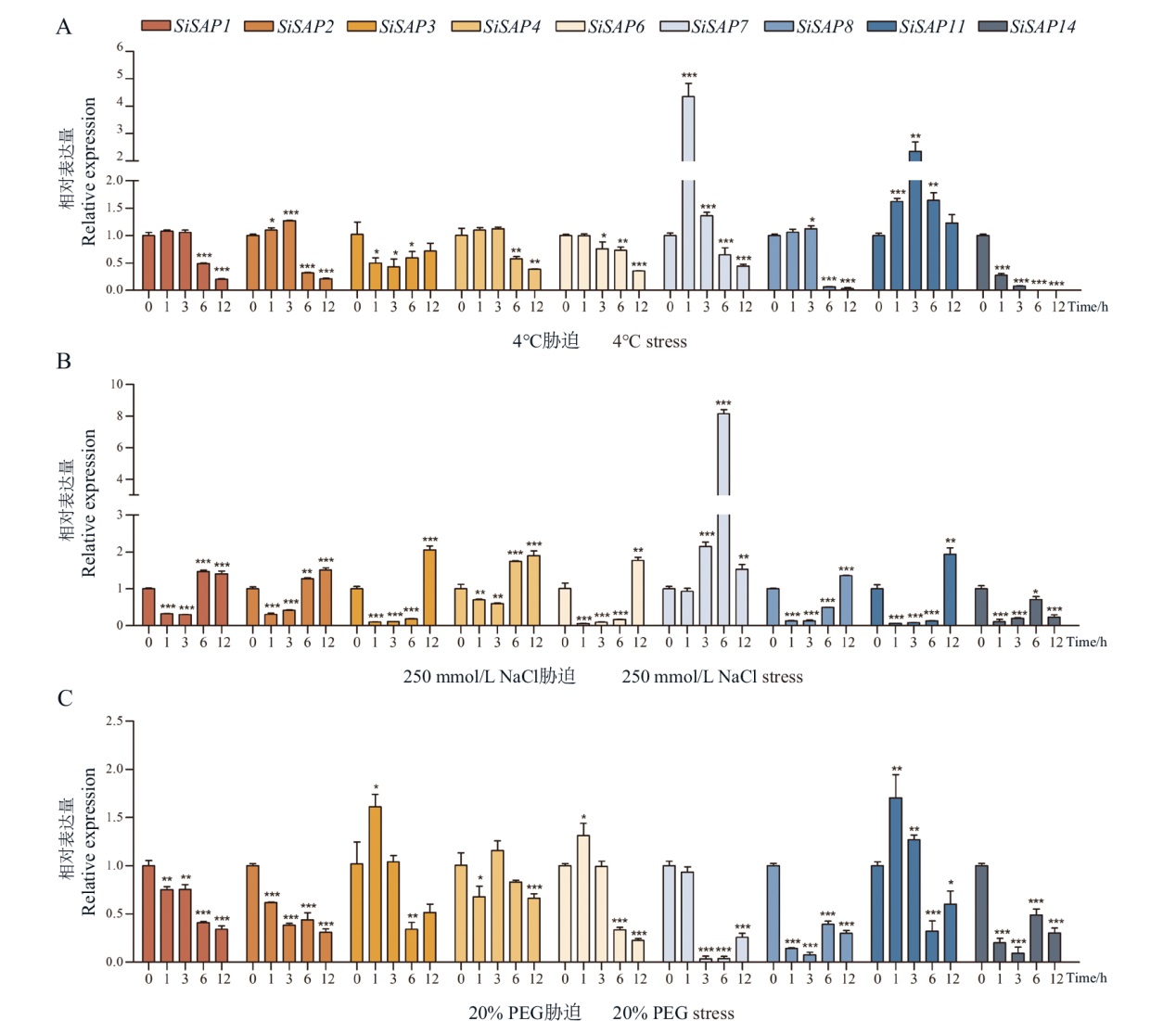
Fig. 9 Expression patterns of SiSAP gene in response to abiotic stress Abscissa indicates 0, 1, 3, 6 and 12 h after treatment. *, **, *** indicate P < 0.05, P < 0.01 and P < 0.001, respectively. The same below
| [1] |
Kanneganti V, Gupta AK. Overexpression of OsiSAP8, a member of stress associated protein(SAP)gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice[J]. Plant Mol Biol, 2008, 66(5): 445-462.
doi: 10.1007/s11103-007-9284-2 pmid: 18205020 |
| [2] | Yoon Y, Seo DH, Shin H, et al. The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants[J]. Agronomy, 2020, 10(6): 788. |
| [3] | Wan FX, Xu YH, Wang SL, et al. Identification and expression analysis of zinc finger A20/AN1 stress-associated genes SmSAP responding to abiotic stress in eggplant[J]. Horticulturae, 2022, 8(2): 108. |
| [4] | Zhao X, Wang R, Zhang Y, et al. Comprehensive analysis of the stress associated protein(SAP)gene family in Tamarix hispida and the function of ThSAP6 in salt tolerance[J]. Plant Physiol Biochem, 2021, 165: 1-9. |
| [5] |
丁兰, 顾宝, 李培楹, 等. 葡萄SAP家族的鉴定与表达分析[J]. 中国农业科学, 2019, 52(14): 2500-2514.
doi: 10.3864/j.issn.0578-1752.2019.14.009 |
| Ding L, Gu B, Li PY, et al. Genome-wide identification and expression analysis of SAP family in grape[J]. Sci Agric Sin, 2019, 52(14): 2500-2514. | |
| [6] |
杜志烨, 李明玉, 陈稷, 等. 植物胁迫相关蛋白功能研究进展[J]. 植物学报, 2024, 59(1): 110-121.
doi: 10.11983/CBB23029 |
| Du ZY, Li MY, Chen J, et al. Research advances in plant stress associated protein functions[J]. Chin Bull Bot, 2024, 59(1): 110-121. | |
| [7] |
Mukhopadhyay A, Vij S, Tyagi AK. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco[J]. Proc Natl Acad Sci USA, 2004, 101(16): 6309-6314.
doi: 10.1073/pnas.0401572101 pmid: 15079051 |
| [8] | 苏晓帅, 张宝华, 刘佳静, 等. 小麦SAPs家族分析及TaSAP1;1耐盐和低磷胁迫功能研究[J]. 植物遗传资源学报, 2022, 23(3): 857-870. |
| Su XS, Zhang BH, Liu JJ, et al. Genomic analysis of stress associated proteins in wheat and functional study of TaSAP1;1 in salt and low-pi tolerance[J]. J Plant Genet Resour, 2022, 23(3): 857-870. | |
| [9] | Vij S, Tyagi AK. Genome-wide analysis of the stress associated protein(SAP)gene family containing A20/AN1 zinc-finger(s)in rice and their phylogenetic relationship with Arabidopsis[J]. Mol Genet Genom, 2006, 276(6): 565-575. |
| [10] |
Baidyussen A, Aldammas M, Kurishbayev A, et al. Identification, gene expression and genetic polymorphism of zinc finger A20/AN1 stress-associated genes, HvSAP, in salt stressed barley from Kazakhstan[J]. BMC Plant Biol, 2020, 20(Suppl 1): 156.
doi: 10.1186/s12870-020-02332-4 pmid: 33050881 |
| [11] | Zhu FJ, Wang K, Li DN, et al. OsSAP6 positively regulates soda saline-alkaline stress tolerance in rice[J]. Rice, 2022, 15(1): 69. |
| [12] | Wang X, Zou BH, Shao QL, et al. Natural variation reveals that OsSAP16 controls low-temperature germination in rice[J]. J Exp Bot, 2018, 69(3): 413-421. |
| [13] |
丁林云, 张微, 王晋成, 等. 过量表达棉花GhSAP1提高转基因烟草的耐盐性[J]. 中国农业科学, 2014, 47(8): 1458-1470.
doi: 10.3864/j.issn.0578-1752.2014.08.002 |
| Ding LY, Zhang W, Wang JC, et al. Overexpression of a Gossypium hirsutum stress-associated protein gene(GhSAP1)improves salt stress tolerance in transgenic tobacco[J]. Sci Agric Sin, 2014, 47(8): 1458-1470. | |
| [14] | 王亦学, 董艳辉, 张欢欢, 等. 陆地棉胁迫相关蛋白基因GhSAP8的克隆及其耐盐性分析[J]. 农业生物技术学报, 2020, 28(7): 1230-1239. |
| Wang YX, Dong YH, Zhang HH, et al. Cloning and salt-tolerance analysis of stress associated protein gene GhSAP8 in Gossypium hirsutum[J]. J Agric Biotechnol, 2020, 28(7): 1230-1239. | |
| [15] |
王亦学, 郝曜山, 张欢欢, 等. 小麦胁迫相关蛋白基因TaSAP12-D的耐盐性分析[J]. 核农学报, 2023, 37(1): 42-50.
doi: 10.11869/j.issn.1000-8551.2023.01.0042 |
| Wang YX, Hao YS, Zhang HH, et al. Identification and analysis of salt-tolerance of stress associated protein gene(TaSAP12-D)from wheat[J]. J Nucl Agric Sci, 2023, 37(1): 42-50. | |
| [16] | Xu QF, Mao XG, Wang YX, et al. A wheat gene TaSAP17-D encoding an AN1/AN1 zinc finger protein improves salt stress tolerance in transgenic Arabidopsis[J]. J Integr Agric, 2018, 17(3): 507-516. |
| [17] | Sharma G, Giri J, Tyagi AK. Rice OsiSAP7 negatively regulates ABA stress signalling and imparts sensitivity to water-deficit stress in Arabidopsis[J]. Plant Sci, 2015, 237: 80-92. |
| [18] | Ben Saad R, Ben Romdhane W, Mihoubi W, et al. A Lobularia maritima LmSAP protein modulates gibberellic acid homeostasis via its A20 domain under abiotic stress conditions[J]. PLoS One, 2020, 15(5): e0233420. |
| [19] |
李顺国, 刘斐, 刘猛, 等. 中国谷子产业和种业发展现状与未来展望[J]. 中国农业科学, 2021, 54(3): 459-470.
doi: 10.3864/j.issn.0578-1752.2021.03.001 |
|
Li SG, Liu F, Liu M, et al. Current status and future prospective of foxtail millet production and seed industry in China[J]. Sci Agric Sin, 2021, 54(3): 459-470.
doi: 10.3864/j.issn.0578-1752.2021.03.001 |
|
| [20] | Lai DL, Fan Y, Xue GX, et al. Genome-wide identification and characterization of the SPL gene family and its expression in the various developmental stages and stress conditions in foxtail millet(Setaria italica)[J]. BMC Genomics, 2022, 23(1): 389. |
| [21] |
Fan Y, Lai DL, Yang H, et al. Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet(Setaria italica L.)[J]. BMC Genomics, 2021, 22(1): 778.
doi: 10.1186/s12864-021-08095-y pmid: 34717536 |
| [22] |
Fan Y, Wei XB, Lai DL, et al. Genome-wide investigation of the GRAS transcription factor family in foxtail millet(Setaria italica L.)[J]. BMC Plant Biol, 2021, 21(1): 508.
doi: 10.1186/s12870-021-03277-y pmid: 34732123 |
| [23] | 雷彪, 曲瑞芳, 任超, 等. 谷子LBD基因家族的鉴定及其对逆境胁迫的响应[J]. 植物生理学报, 2023, 59(3): 527-542. |
| Lei B, Qu RF, Ren C, et al. Identification of SiLBDs gene family in foxtail millet and its participation in stress response[J]. Plant Physiol J, 2023, 59(3): 527-542. | |
| [24] | 桑璐曼, 汤沙, 张仁梁, 等. 谷子热激蛋白HSP90基因家族鉴定及分析[J]. 植物遗传资源学报, 2022, 23(4): 1085-1097. |
| Sang LM, Tang S, Zhang RL, et al. Identification and analysis of heat shock protein HSP90 family genes in foxtail millet[J]. J Plant Genet Resour, 2022, 23(4): 1085-1097. | |
| [25] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining[J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [26] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [27] |
Jeffares DC, Penkett CJ, Bähler J. Rapidly regulated genes are intron poor[J]. Trends Genet, 2008, 24(8): 375-378.
doi: 10.1016/j.tig.2008.05.006 pmid: 18586348 |
| [28] |
Giri J, Dansana PK, Kothari KS, et al. SAPs as novel regulators of abiotic stress response in plants[J]. Bioessays, 2013, 35(7): 639-648.
doi: 10.1002/bies.201200181 pmid: 23640876 |
| [29] | Gao W, Long L, Tian XQ, et al. Genome-wide identification and expression analysis of stress-associated proteins(SAPs)containing A20/AN1 zinc finger in cotton[J]. Mol Genet Genomics, 2016, 291(6): 2199-2213. |
| [30] | Zhou Y, Zeng LM, Chen RR, et al. Genome-wide identification and characterization of stress-associated protein(SAP)gene family encoding A20/AN1 zinc-finger proteins in Medicago truncatula[J]. Arch Biol Sci, 2018, 70(1): 87-98. |
| [31] | Dong QL, Duan DY, Zhao S, et al. Genome-wide analysis and cloning of the apple stress-associated protein gene family reveals MdSAP15, which confers tolerance to drought and osmotic stresses in transgenic Arabidopsis[J]. Int J Mol Sci, 2018, 19(9): 2478. |
| [32] | Kong FL, Wang J, Cheng L, et al. Genome-wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum[J]. Gene, 2012, 499(1): 108-120. |
| [33] |
Matsukura S, Mizoi J, Yoshida T, et al. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes[J]. Mol Genet Genomics, 2010, 283(2): 185-196.
doi: 10.1007/s00438-009-0506-y pmid: 20049613 |
| [34] |
Tyagi H, Jha S, Sharma M, et al. Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco[J]. Plant Sci, 2014, 225: 68-76.
doi: 10.1016/j.plantsci.2014.05.016 pmid: 25017161 |
| [35] | Kothari KS, Dansana PK, Giri J, et al. Rice stress associated protein 1(OsSAP1)interacts with aminotransferase(OsAMTR1)and pathogenesis-related 1a protein(OsSCP)and regulates abiotic stress responses[J]. Front Plant Sci, 2016, 7: 1057. |
| [36] | Muthuramalingam P, Jeyasri R, Selvaraj A, et al. Global transcriptome analysis of novel stress associated protein(SAP)genes expression dynamism of combined abiotic stresses in Oryza sativa(L.)[J]. J Biomol Struct Dyn, 2021, 39(6): 2106-2117. |
| [1] | YIN Yuan, CHENG Shuang, LIU Ding-hao, DENG Xiao-xia, LI Kai-yue, WANG Jing-hong, LIN Ji-xiang. Research Progress in Exogenous Hydrogen Peroxide(H2O2)Affecting Plant Growth and Physiological Metabolism under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(1): 1-13. |
| [2] | WU Zhi-jian, LIU Guang-yang, LIN Zhi-hao, SHENG Bin, CHEN Ge, XU Xiao-min, WANG Jun-wei, XU Dong-hui. Research Progress of Nano-regulation of Vegetable Seed Germination and Its Mechanism [J]. Biotechnology Bulletin, 2025, 41(1): 14-24. |
| [3] | KONG Qing-yang, ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le. Identification and Expression Analysis of GRAS Transcription Factor Family in Rosa persica [J]. Biotechnology Bulletin, 2025, 41(1): 210-220. |
| [4] | WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper [J]. Biotechnology Bulletin, 2024, 40(9): 198-211. |
| [5] | MAN Quan-cai, MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui. Identification and Expression Analysis of AQP Gene Family in Potato [J]. Biotechnology Bulletin, 2024, 40(9): 51-63. |
| [6] | SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers [J]. Biotechnology Bulletin, 2024, 40(9): 104-112. |
| [7] | SHEN Peng, GAO Ya-Bin, DING Hong. Identification and Expression Analysis of SAT Gene Family in Potato(Solanum tuberosum L.) [J]. Biotechnology Bulletin, 2024, 40(9): 64-73. |
| [8] | LI Yi-jun, YANG Xiao-bei, XIA Lin, LUO Zhao-peng, XU Xin, YANG Jun, NING Qian-ji, WU Ming-zhu. Cloning and Functional Analysis of NtPRR37 Gene in Nicotiana tabacum L. [J]. Biotechnology Bulletin, 2024, 40(8): 221-231. |
| [9] | LI Yong-hui, BAO Xing-xing, DUAN Yi-ke, ZHAO Yun-xia, YU Xiang-li, CHEN Yao, ZHANG Yan-zhao. Genome-wide Identification and Expression Features Analysis of the bZIP Family in Rhododendron henanense subsp. lingbaoense [J]. Biotechnology Bulletin, 2024, 40(8): 186-198. |
| [10] | ZHOU Ran, WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma. Analysis of LncRNA Differential Expression in Mammary Tissue of Cows with Staphylococcus aureus Mastitis [J]. Biotechnology Bulletin, 2024, 40(8): 320-328. |
| [11] | LI Yu-qing, WU Nan, LUO Jian-rang. Cloning and Functional Analysis of bHLH Gene Related to Anthocyanin Synthesis in Paeonia qiui [J]. Biotechnology Bulletin, 2024, 40(8): 174-185. |
| [12] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [13] | LIU Dan-dan, WANG Lei-gang, SUN Ming-hui, JIAO Xiao-yu, WU Qiong, WANG Wen-jie. Genome-wide Identification and Expression Pattern Profiling of the Trehalose-6-phosphate Synthase(TPS)Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2024, 40(8): 152-163. |
| [14] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [15] | YU Niu, LIU Fan, YANG Jin-chang. Cloning of SgTPS7 in Sindora glabra and Its Function in Terpene Synthesis and Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(8): 164-173. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||