








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 152-163.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0252
Previous Articles Next Articles
LIU Dan-dan( ), WANG Lei-gang, SUN Ming-hui, JIAO Xiao-yu, WU Qiong, WANG Wen-jie(
), WANG Lei-gang, SUN Ming-hui, JIAO Xiao-yu, WU Qiong, WANG Wen-jie( )
)
Received:2024-03-15
Online:2024-08-26
Published:2024-07-30
Contact:
WANG Wen-jie
E-mail:1653082943@qq.com;wwj00@126.com
LIU Dan-dan, WANG Lei-gang, SUN Ming-hui, JIAO Xiao-yu, WU Qiong, WANG Wen-jie. Genome-wide Identification and Expression Pattern Profiling of the Trehalose-6-phosphate Synthase(TPS)Gene Family in Tea Plant(Camellia sinensis)[J]. Biotechnology Bulletin, 2024, 40(8): 152-163.
| 基因名称Gene name | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|
| β-actin | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT |
| CsTPS3 | TTCTCACAGAGAGTGATTGAGGTG | GGACTATGGAGAAAGAACCCCATT |
| CsTPS5 | TTCTCACAGAGAGTGATTGAGGTG | GGACTATGGAGAAAGAACCCCATT |
| CsTPS6 | TGTCTTTATTGTTAGTGGGAGGGG | CACAATTTCCTTCCACTCAAGGTC |
| CsTPS9 | GCAAGATATGGAGAGAACTTGTGC | TGGGGCATTACAGTACCATCATAG |
| CsTPS11 | GGTTACCTGTCTCTGCAGTTAGAA | CAAAGCTTTAGTAAGTGCCCTCTG |
| CsTPS14 | GCCCTGATGATGATTTTGTATGGG | TTCAACTCTGACAGGTAGTGTACG |
Table 1 Primer sequences for real-time PCR
| 基因名称Gene name | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|
| β-actin | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT |
| CsTPS3 | TTCTCACAGAGAGTGATTGAGGTG | GGACTATGGAGAAAGAACCCCATT |
| CsTPS5 | TTCTCACAGAGAGTGATTGAGGTG | GGACTATGGAGAAAGAACCCCATT |
| CsTPS6 | TGTCTTTATTGTTAGTGGGAGGGG | CACAATTTCCTTCCACTCAAGGTC |
| CsTPS9 | GCAAGATATGGAGAGAACTTGTGC | TGGGGCATTACAGTACCATCATAG |
| CsTPS11 | GGTTACCTGTCTCTGCAGTTAGAA | CAAAGCTTTAGTAAGTGCCCTCTG |
| CsTPS14 | GCCCTGATGATGATTTTGTATGGG | TTCAACTCTGACAGGTAGTGTACG |
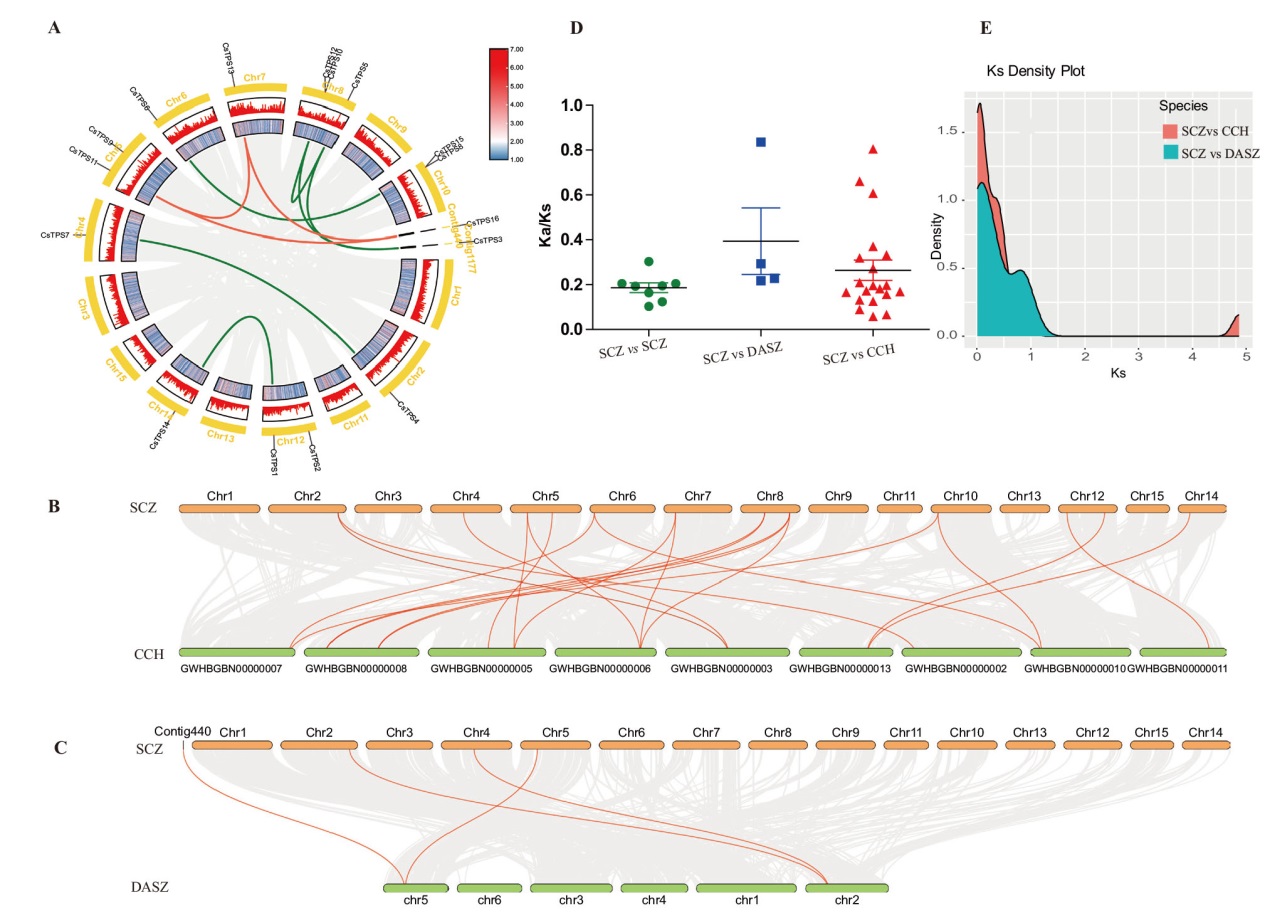
Fig. 5 Analysis of syntenic and Ka/Ks of TPS genes in Camellia plants A: Syntenic analysis of TPS genes in C. sinensis. The inner gray lines indicate all the collinearity relationships in the whole genome, and the inner orange and green lines indicate the collinearity relationships of Class I and Class II subfamilies, respectively. B: Syntenic analysis of TPS genes between C. sinensis ‘SCZ’ and C. chekiangoleosa. C: Syntenic analysis of TPS genes between C. sinensis ‘SCZ’ and C. sinensis ‘DASZ’. D: Analysis of Ka/Ks. E: Density distribution of Ks for paralogous gene pairs of the three species of Camellia plants
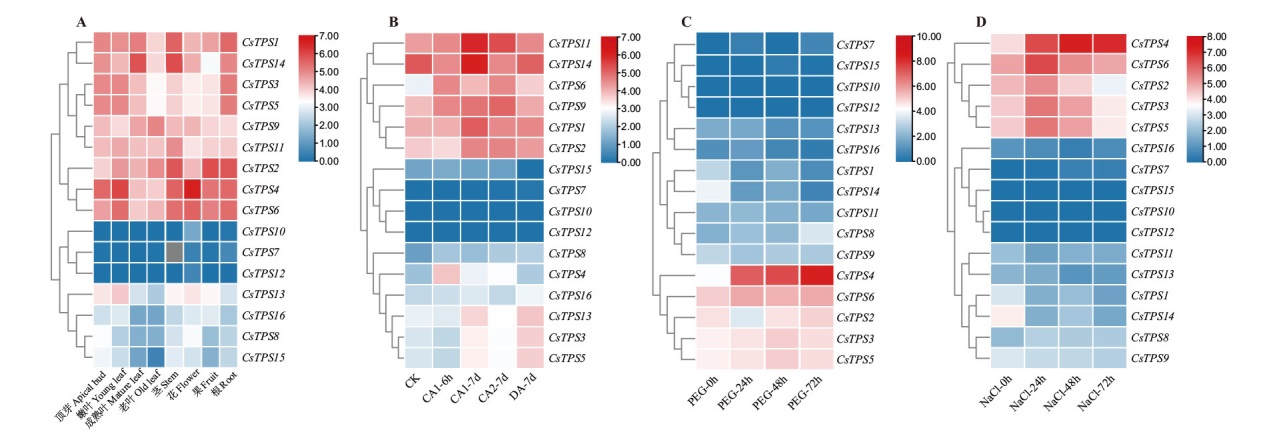
Fig. 6 Transcriptome expression profiling of TPS genes in C. sinensis A, C and D graphs indicate the expression patterns of CsTPS genes in different tissues, 25% PEG stress and 200 mmol/L NaCl stress, respectively. B: Expression pattern of CsTPS gene under low temperature domestication(CK: 1 month of growth at 25℃ during the day, 20℃ at night, 12 h photoperiod and 70% humidity; CA1-6h: 6 h treatment at 10℃; CA1-7d: 10℃ during the day, 4℃ at night treatment for 7 d; CA2-7d: 4℃ during the day and 0℃ at night for 7 d; DA-7d: 25℃ during the day and 20℃ at night for 7 d). The log2(FPKM+1)value reflects in the heatmap
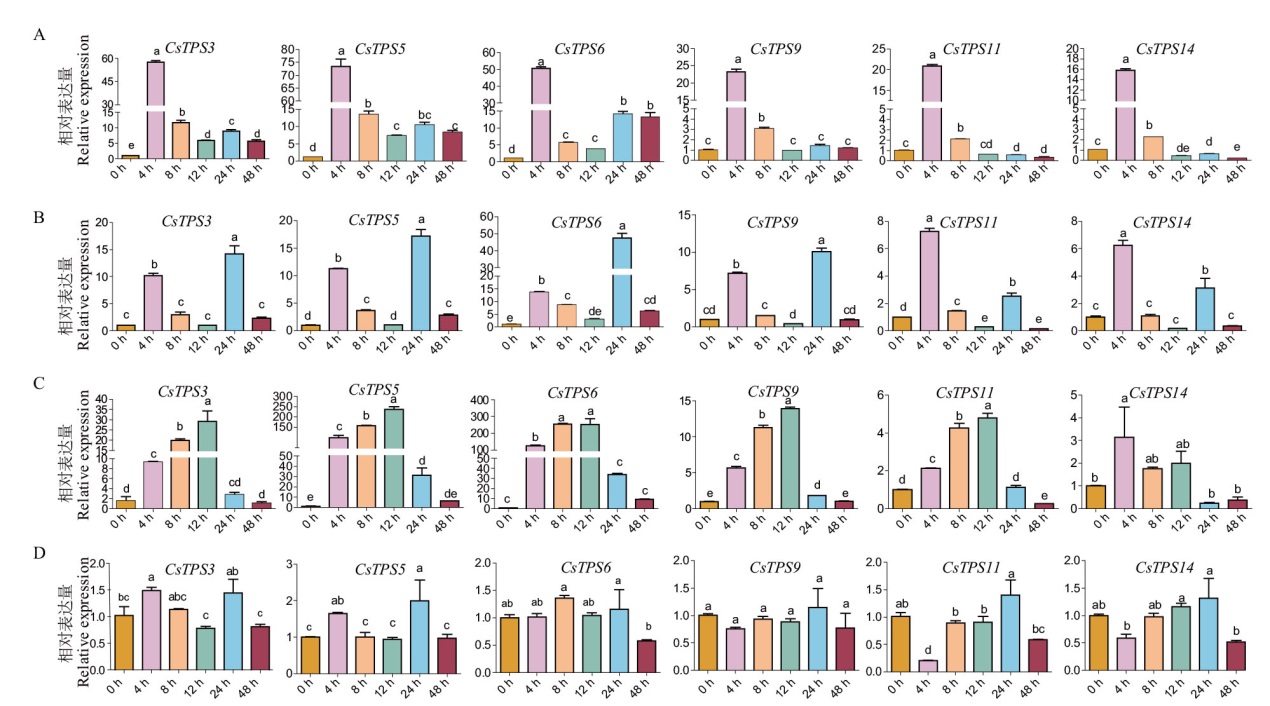
Fig. 7 Relative expressions of CsTPS genes under various stress A: Gene expression profiles of CsTPSs under 20% PEG. B: Gene expression profiles of CsTPSs under 200 mmol/L NaCl. C: Gene expression profiles of CsTPSs under 4℃ treatment. D: Circadian expression profiling of the CsTPS genes at 24℃. Error bars indicate the standard deviation of three independent biological replicates, different lowercase letters in indicate significant differences at different time of the same treatment(P<0.05)
| [1] | Liu Y, Lee J, Mansfield KM, et al. Trehalose glycopolymer enhances both solution stability and pharmacokinetics of a therapeutic protein[J]. Bioconjug Chem, 2017, 28(3): 836-845. |
| [2] | Acosta-Pérez P, Camacho-Zamora BD, Espinoza-Sánchez EA, et al. Characterization of trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase genes and analysis of its differential expression in maize(Zea mays)seedlings under drought stress[J]. Plants, 2020, 9(3): 315. |
| [3] |
Márquez-Escalante JA, Figueroa-Soto CG, Valenzuela-Soto EM. Isolation and partial characterization of trehalose 6-phosphate synthase aggregates from Selaginella lepidophylla plants[J]. Biochimie, 2006, 88(10): 1505-1510.
pmid: 16828951 |
| [4] | Fichtner F, Lunn JE. The role of trehalose 6-phosphate(Tre6P)in plant metabolism and development[J]. Annu Rev Plant Biol, 2021, 72: 737-760. |
| [5] |
Vogel G, Aeschbacher RA, Müller J, et al. Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: identification by functional complementation of the yeast tps2 mutant[J]. Plant J, 1998, 13(5): 673-683.
doi: 10.1046/j.1365-313x.1998.00064.x pmid: 9681009 |
| [6] |
Blázquez MA, Santos E, Flores CL, et al. Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase[J]. Plant J, 1998, 13(5): 685-689.
doi: 10.1046/j.1365-313x.1998.00063.x pmid: 9681010 |
| [7] |
Vandesteene L, López-Galvis L, Vanneste K, et al. Expansive evolution of the trehalose-6-phosphate phosphatase gene family in Arabidopsis[J]. Plant Physiol, 2012, 160(2): 884-896.
doi: 10.1104/pp.112.201400 pmid: 22855938 |
| [8] |
Ponnu J, Wahl V, Schmid M. Trehalose-6-phosphate: connecting plant metabolism and development[J]. Front Plant Sci, 2011, 2: 70.
doi: 10.3389/fpls.2011.00070 pmid: 22639606 |
| [9] | Figueroa CM, Feil R, Ishihara H, et al. Trehalose 6-phosphate coordinates organic and amino acid metabolism with carbon availability[J]. Plant J, 2016, 85(3): 410-423. |
| [10] | Lunn JE, Delorge I, Figueroa CM, et al. Trehalose metabolism in plants[J]. Plant J, 2014, 79(4): 544-567. |
| [11] | Avonce N, Wuyts J, Verschooten K, et al. The Cytophagahutchin-soniiChTPSP: first characterized bifunctional TPS-TPP protein as putative ancestor of all eukaryotic trehalose biosynthesis proteins[J]. Mol Biol Evol, 2010, 27(2): 359-369. |
| [12] |
Paul MJ, Primavesi LF, Jhurreea D, et al. Trehalose metabolism and signaling[J]. Annu Rev Plant Biol, 2008, 59: 417-441.
doi: 10.1146/annurev.arplant.59.032607.092945 pmid: 18257709 |
| [13] |
Miranda JA, Avonce N, Suárez R, et al. A bifunctional TPS-TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis[J]. Planta, 2007, 226(6): 1411-1421.
pmid: 17628825 |
| [14] | Chary SN, Hicks GR, Choi YG, et al. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis[J]. Plant Physiol, 2008, 146(1): 97-107. |
| [15] | Satoh-Nagasawa N, Nagasawa N, Malcomber S, et al. A trehalose metabolic enzyme controls inflorescence architecture in maize[J]. Nature, 2006, 441(7090): 227-230. |
| [16] |
Eastmond PJ, Spielman M, et al. Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation[J]. Plant J, 2002, 29(2): 225-235.
doi: 10.1046/j.1365-313x.2002.01220.x pmid: 11851922 |
| [17] | Gómez LD, Gilday A, Feil R, et al. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells[J]. Plant J, 2010, 64(1): 1-13. |
| [18] | Schluepmann H, Smeekens SCM. Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering[J]. Plant Physiol, 2004, 135(2): 969-977. |
| [19] |
Vandesteene L, Ramon M, Le Roy K, et al. A single active trehalose-6-P synthase(TPS)and a family of putative regulatory TPS-like proteins in Arabidopsis[J]. Mol Plant, 2010, 3(2): 406-419.
doi: 10.1093/mp/ssp114 pmid: 20100798 |
| [20] | Zang BS, Li HW, Li WJ, et al. Analysis of trehalose-6-phosphate synthase(TPS)gene family suggests the formation of TPS complexes in rice[J]. Plant Mol Biol, 2011, 76(6): 507-522. |
| [21] | Gao YH, Yang XY, Yang X, et al. Characterization and expression pattern of the trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase gene families in Populus[J]. Int J Biol Macromol, 2021, 187: 9-23. |
| [22] | Xu YC, Wang YJ, Mattson N, et al. Genome-wide analysis of the Solanum tuberosum(potato)trehalose-6-phosphate synthase(TPS)gene family: evolution and differential expression during development and stress[J]. BMC Genomics, 2017, 18(1): 926. |
| [23] | Du LS, Qi SY, Ma JJ, et al. Identification of TPS family members in apple(Malus×domestica Borkh.)and the effect of sucrose sprays on TPS expression and floral induction[J]. Plant PhysiolBiochem, 2017, 120: 10-23. |
| [24] | Dan YY, Niu Y, Wang CL, et al. Genome-wide identification and expression analysis of the trehalose-6-phosphate synthase(TPS)gene family in cucumber(Cucumis sativus L.)[J]. PeerJ, 2021, 9: e11398. |
| [25] |
Delorge I, Janiak M, Carpentier S, et al. Fine tuning of trehalose biosynthesis and hydrolysis as novel tools for the generation of abiotic stress tolerant plants[J]. Front Plant Sci, 2014, 5: 147.
doi: 10.3389/fpls.2014.00147 pmid: 24782885 |
| [26] | Ramon M, De Smet I, Vandesteene L, et al. Extensive expression regulation and lack of heterologous enzymatic activity of the Class II trehalose metabolism proteins from Arabidopsis thaliana[J]. Plant Cell Environ, 2009, 32(8): 1015-1032. |
| [27] | Avonce N, Leyman B, Mascorro-Gallardo JO, et al. The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling[J]. Plant Physiol, 2004, 136(3): 3649-3659. |
| [28] | Almeida AM, Villalobos E, Araújo SS, et al. Transformation of tobacco with an Arabidopsis thaliana gene involved in trehalose biosynthesis increases tolerance to several abiotic stresses[J]. Euphytica, 2005, 146(1): 165-176. |
| [29] | Li HW, Zang BS, Deng XW, et al. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice[J]. Planta, 2011, 234(5): 1007-1018. |
| [30] | Tian LF, Xie ZJ, Lu CQ, et al. The trehalose-6-phosphate synthase TPS5 negatively regulates ABA signaling in Arabidopsis thaliana[J]. Plant Cell Rep, 2019, 38(8): 869-882. |
| [31] | 丁菲, 庞磊, 李叶云, 等. 茶树海藻糖-6-磷酸合成酶基因(CsTPS)的克隆及表达分析[J]. 农业生物技术学报, 2012, 20(11): 1253-1261. |
| Ding F, Pang L, Li YY, et al. Cloning and expression analysis of trehalose-6-phosphate synthase gene(CsTPS)from tea plant(Camellia sinensis(L.) O. kuntz)[J]. J Agric Biotechnol, 2012, 20(11): 1253-1261. | |
| [32] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining[J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [33] | Wei CL, Yang H, Wang SB, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality[J]. Proc Natl Acad Sci USA, 2018, 115(18): E4151-E4158. |
| [34] | Zhang Q, Cai MC, Yu XM, et al. Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress[J]. Tree Genet Genomes, 2017, 13(4): 78. |
| [35] | Li YY, Wang XW, Ban QY, et al. Comparative transcriptomic analysis reveals gene expression associated with cold adaptation in the tea plant Camellia sinensis[J]. BMC Genomics, 2019, 20(1): 624. |
| [36] | Cao QH, Lv WY, Jiang H, et al. Genome-wide identification of glutathione S-transferase gene family members in tea plant(Camellia sinensis)and their response to environmental stress[J]. Int J Biol Macromol, 2022, 205: 749-760. |
| [37] | Li PH, Xia EH, Fu JM, et al. Diverse roles of MYB transcription factors in regulating secondary metabolite biosynthesis, shoot development, and stress responses in tea plants(Camellia sinensis)[J]. Plant J, 2022, 110(4): 1144-1165. |
| [38] |
Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution[J]. Trends Genet, 2002, 18(9): 486.
doi: 10.1016/s0168-9525(02)02722-1 pmid: 12175810 |
| [39] | Song J, Gao ZH, Huo XM, et al. Genome-wide identification of the auxin response factor(ARF)gene family and expression analysis of its role associated with pistil development in Japanese apricot(Prunus mume Sieb. et Zucc)[J]. Acta Physiol Plant, 2015, 37(8): 145. |
| [40] | Xia EH, Zhang HB, Sheng J, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis[J]. Mol Plant, 2017, 10(6): 866-877. |
| [41] | Shen TF, Huang B, Xu M, et al. The reference genome of Camellia chekiangoleosa provides insights into camellia evolution and tea oil biosynthesis[J]. Hortic Res, 2022, 9: uhab083. |
| [42] | Doi K, Hosaka A, Nagata T, et al. Development of a novel data mining tool to find cis-elements in rice gene promoter regions[J]. BMC Plant Biol, 2008, 8(1): 20. |
| [43] |
Gómez-Porras JL, Riaño-Pachón DM, Dreyer I, et al. Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice[J]. BMC Genomics, 2007, 8: 260.
pmid: 17672917 |
| [44] |
Karim S, Aronsson H, Ericson H, et al. Improved drought tolerance without undesired side effects in transgenic plants producing trehalose[J]. Plant Mol Biol, 2007, 64(4): 371-386.
doi: 10.1007/s11103-007-9159-6 pmid: 17453154 |
| [45] |
Garg AK, Kim JK, Owens TG, et al. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses[J]. Proc Natl Acad Sci USA, 2002, 99(25): 15898-15903.
pmid: 12456878 |
| [46] | Jang IC, Oh SJ, Seo JS, et al. Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth[J]. Plant Physiol, 2003, 131(2): 516-524. |
| [47] |
Romero C, Bellés JM, Vayá JL, et al. Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance[J]. Planta, 1997, 201(3): 293-297.
doi: 10.1007/s004250050069 pmid: 19343407 |
| [48] |
Vishal B, Krishnamurthy P, Ramamoorthy R, et al. OsTPS8 controls yield-related traits and confers salt stress tolerance in rice by enhancing suberin deposition[J]. New Phytol, 2019, 221(3): 1369-1386.
doi: 10.1111/nph.15464 pmid: 30289560 |
| [49] | Veyres N, Danon A, Aono M, et al. The Arabidopsis sweetie mutant is affected in carbohydrate metabolism and defective in the control of growth, development and senescence[J]. Plant J, 2008, 55(4): 665-686. |
| [50] |
Fernandez O, Béthencourt L, Quero A, et al. Trehalose and plant stress responses: friend or foe?[J]. Trends Plant Sci, 2010, 15(7): 409-417.
doi: 10.1016/j.tplants.2010.04.004 pmid: 20494608 |
| [51] | Lunn JE, Feil R, Hendriks JHM, et al. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucosepyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana[J]. Biochem J, 2006, 397(1): 139-148. |
| [52] |
Nunes C, O'Hara LE, Primavesi LF, et al. The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation[J]. Plant Physiol, 2013, 162(3): 1720-1732.
doi: 10.1104/pp.113.220657 pmid: 23735508 |
| [1] | LI Yong-hui, BAO Xing-xing, DUAN Yi-ke, ZHAO Yun-xia, YU Xiang-li, CHEN Yao, ZHANG Yan-zhao. Genome-wide Identification and Expression Features Analysis of the bZIP Family in Rhododendron henanense subsp. lingbaoense [J]. Biotechnology Bulletin, 2024, 40(8): 186-198. |
| [2] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [3] | YU Niu, LIU Fan, YANG Jin-chang. Cloning of SgTPS7 in Sindora glabra and Its Function in Terpene Synthesis and Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(8): 164-173. |
| [4] | WU Ding-jie, CHEN Ying-ying, XU Jing, LIU Yuan, ZHANG Hang, LI Rui-li. Research Progress in Plant Gibberellin Oxidase and Its Functions [J]. Biotechnology Bulletin, 2024, 40(7): 43-54. |
| [5] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [6] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [7] | DU Bing-shuai, ZOU Xin-hui, WANG Zi-hao, ZHANG Xin-yuan, CAO Yi-bo, ZHANG Ling-yun. Genome-wide Identification and Expression Analysis of the SWEET Gene Family in Camellia oleifera [J]. Biotechnology Bulletin, 2024, 40(5): 179-190. |
| [8] | GUO Hui-yan, DONG Xue, AN Meng-nan, XIA Zi-hao, WU Yuan-hua. Research Progress in the Functions of Key Enzymes of Ubiquitination Modification in Plant Stress Responses [J]. Biotechnology Bulletin, 2024, 40(4): 1-11. |
| [9] | JIANG Lin-qi, ZHAO Jia-ying, ZHENG Fei-xiong, YAO Xin-yi, LI Xiao-xian, YU Zhen-ming. Identification and Expression Analysis of 14-3-3 Gene Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2024, 40(3): 229-241. |
| [10] | ZHOU Hong-dan, LUO Xiao-ping, TU Mi-xue, LI Zhong-guang. Phytomelatonin: An Emerging Signal Molecule Responding to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(3): 41-51. |
| [11] | WU Cui-cui, XIAO Shui-ping. Genome-wide Identification of HD-Zip Gene Family in Gossypium hirsutum L. and Expression Analysis in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(2): 130-145. |
| [12] | XIN Qi, LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua. Identification and Expression Analysis of the CBL-CIPK Gene Family in Sugarcane [J]. Biotechnology Bulletin, 2024, 40(2): 197-211. |
| [13] | YANG Yu-qing, TAN Juan, WANG Fang, PENG Shun-li, CHEN Jie, TAN Ming-yan, LYU Mei-yan, ZHOU Fu-yu, LIU Sheng-chuan. Research and Application Progress in Chloroplast Genome of Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2024, 40(2): 20-30. |
| [14] | LI Ya-nan, ZHANG Hao-jie, LIANG Meng-jing, LUO Tao, LI Wang-ning, ZHANG Chun-hui, JI Chun-li, LI Run-zhi, XUE Jin-ai, CUI Hong-li. Identification and Expression Analysis of Calcium-dependent Protein Kinase(CDPK)Family in Haematococcus pluvialis [J]. Biotechnology Bulletin, 2024, 40(2): 300-312. |
| [15] | ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses [J]. Biotechnology Bulletin, 2024, 40(2): 90-98. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||