








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (7): 193-204.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1277
Previous Articles Next Articles
WANG Fang1,2( ), QIAO Shuai1,2, SONG Wei1,2, CUI Peng-juan3, LIAO An-zhong1,2, TAN Wen-fang1,2(
), QIAO Shuai1,2, SONG Wei1,2, CUI Peng-juan3, LIAO An-zhong1,2, TAN Wen-fang1,2( ), YANG Song-tao1,2(
), YANG Song-tao1,2( )
)
Received:2024-12-30
Online:2025-07-26
Published:2025-07-22
Contact:
TAN Wen-fang, YANG Song-tao
E-mail:wangfangsaas@126.com;zwstwf414@163.com;yost60@126.com
WANG Fang, QIAO Shuai, SONG Wei, CUI Peng-juan, LIAO An-zhong, TAN Wen-fang, YANG Song-tao. Genome-wide Identification of the IbNRT2 Gene Family and Its Expression in Sweet Potato[J]. Biotechnology Bulletin, 2025, 41(7): 193-204.
| Gene | Forward sequences (5´-3´) | Reverse sequences (5´-3´) |
|---|---|---|
| IbNRT2.1 | TCATGTTAACTGCCCCCACC | AAGAGACGAAAGTGGCGAGG |
| IbNRT2.2 | GGGATGGCTTCATGTGGTGA | GTGACCGCGTACCAAAACAC |
| IbNRT2.3 | GGGTTCCCAAACTATTCCC | AAGGTGGAGATGAAGCAGG |
| IbNRT2.4 | TGGCTGGGATGGAAGATTC | TCTGAGTCAACGGGCAATG |
| IbNRT2.5 | GCGGCTAGTTTTGGATTGGC | CACAGGCAGCTTGAACGAAC |
| IbNRT2.6 | GCCTTCCTCAACCTCCTCAC | GTCCAGAAGGAGCAGGACAC |
| IbNRT2.7 | AATCTCCCAACCCAAGCAGG | CAGAACATTCCGCCCCATCT |
| IbACTIN | GTATTGTGCTGGATTCTGGTGAT | TCAGCAGTAGTGGTGAACATGT |
Table 1 Primer sequences for RT-qPCR
| Gene | Forward sequences (5´-3´) | Reverse sequences (5´-3´) |
|---|---|---|
| IbNRT2.1 | TCATGTTAACTGCCCCCACC | AAGAGACGAAAGTGGCGAGG |
| IbNRT2.2 | GGGATGGCTTCATGTGGTGA | GTGACCGCGTACCAAAACAC |
| IbNRT2.3 | GGGTTCCCAAACTATTCCC | AAGGTGGAGATGAAGCAGG |
| IbNRT2.4 | TGGCTGGGATGGAAGATTC | TCTGAGTCAACGGGCAATG |
| IbNRT2.5 | GCGGCTAGTTTTGGATTGGC | CACAGGCAGCTTGAACGAAC |
| IbNRT2.6 | GCCTTCCTCAACCTCCTCAC | GTCCAGAAGGAGCAGGACAC |
| IbNRT2.7 | AATCTCCCAACCCAAGCAGG | CAGAACATTCCGCCCCATCT |
| IbACTIN | GTATTGTGCTGGATTCTGGTGAT | TCAGCAGTAGTGGTGAACATGT |
| Gene ID | Gene name | Chr | AA Number | Mw | pI | GRAVY | Predicted location |
|---|---|---|---|---|---|---|---|
| IB12G06570 | IbNRT2.1 | 12 | 525 | 56 854.18 | 9.19 | 0.387 | Cell membrane |
| IB12G06560 | IbNRT2.2 | 12 | 524 | 56 885.85 | 8.94 | 0.393 | Cell membrane |
| IB02G04080 | IbNRT2.3 | 2 | 536 | 58 419.54 | 8.90 | 0.382 | Cell membrane |
| IB06G26510 | IbNRT2.4 | 6 | 462 | 50 169.9 | 7.89 | 0.64 | Cell membrane |
| IB10G16710 | IbNRT2.5 | 10 | 502 | 54 245.02 | 9.17 | 0.423 | Cell membrane |
| IB12G06540 | IbNRT2.6 | 12 | 334 | 36 679.66 | 9.41 | 0.25 | Cell membrane |
| IB07G31450 | IbNRT2.7 | 7 | 459 | 50 338.97 | 8.90 | 0.053 | Cell membrane |
Table 2 Analysis on the physical and chemical properties of nitrate transporter IbNRT2 in sweet potato
| Gene ID | Gene name | Chr | AA Number | Mw | pI | GRAVY | Predicted location |
|---|---|---|---|---|---|---|---|
| IB12G06570 | IbNRT2.1 | 12 | 525 | 56 854.18 | 9.19 | 0.387 | Cell membrane |
| IB12G06560 | IbNRT2.2 | 12 | 524 | 56 885.85 | 8.94 | 0.393 | Cell membrane |
| IB02G04080 | IbNRT2.3 | 2 | 536 | 58 419.54 | 8.90 | 0.382 | Cell membrane |
| IB06G26510 | IbNRT2.4 | 6 | 462 | 50 169.9 | 7.89 | 0.64 | Cell membrane |
| IB10G16710 | IbNRT2.5 | 10 | 502 | 54 245.02 | 9.17 | 0.423 | Cell membrane |
| IB12G06540 | IbNRT2.6 | 12 | 334 | 36 679.66 | 9.41 | 0.25 | Cell membrane |
| IB07G31450 | IbNRT2.7 | 7 | 459 | 50 338.97 | 8.90 | 0.053 | Cell membrane |
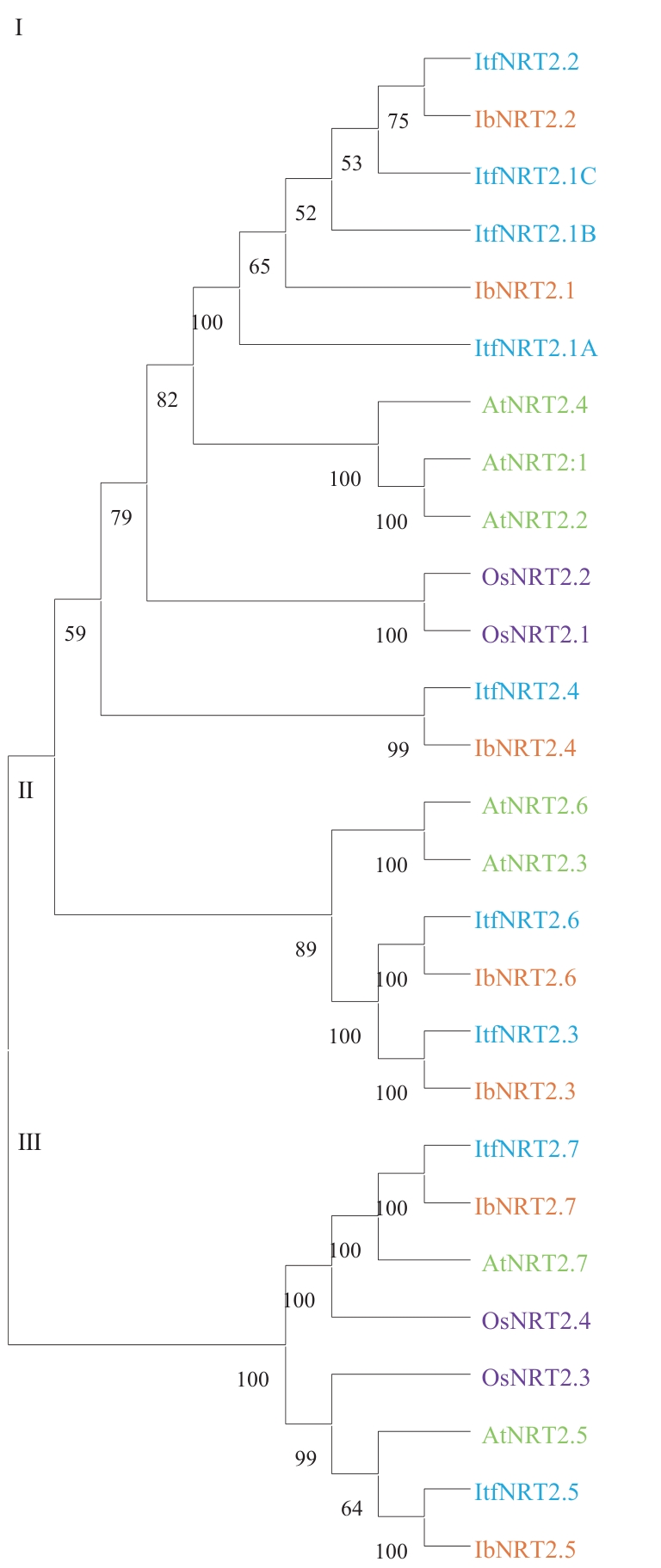
Fig. 3 Phylogenetic tree analysis of NRT2 family member in sweet potato (Ib), closely related wild diploid (Itf), Arabidopsis (At), and Oryza sativa (Os)
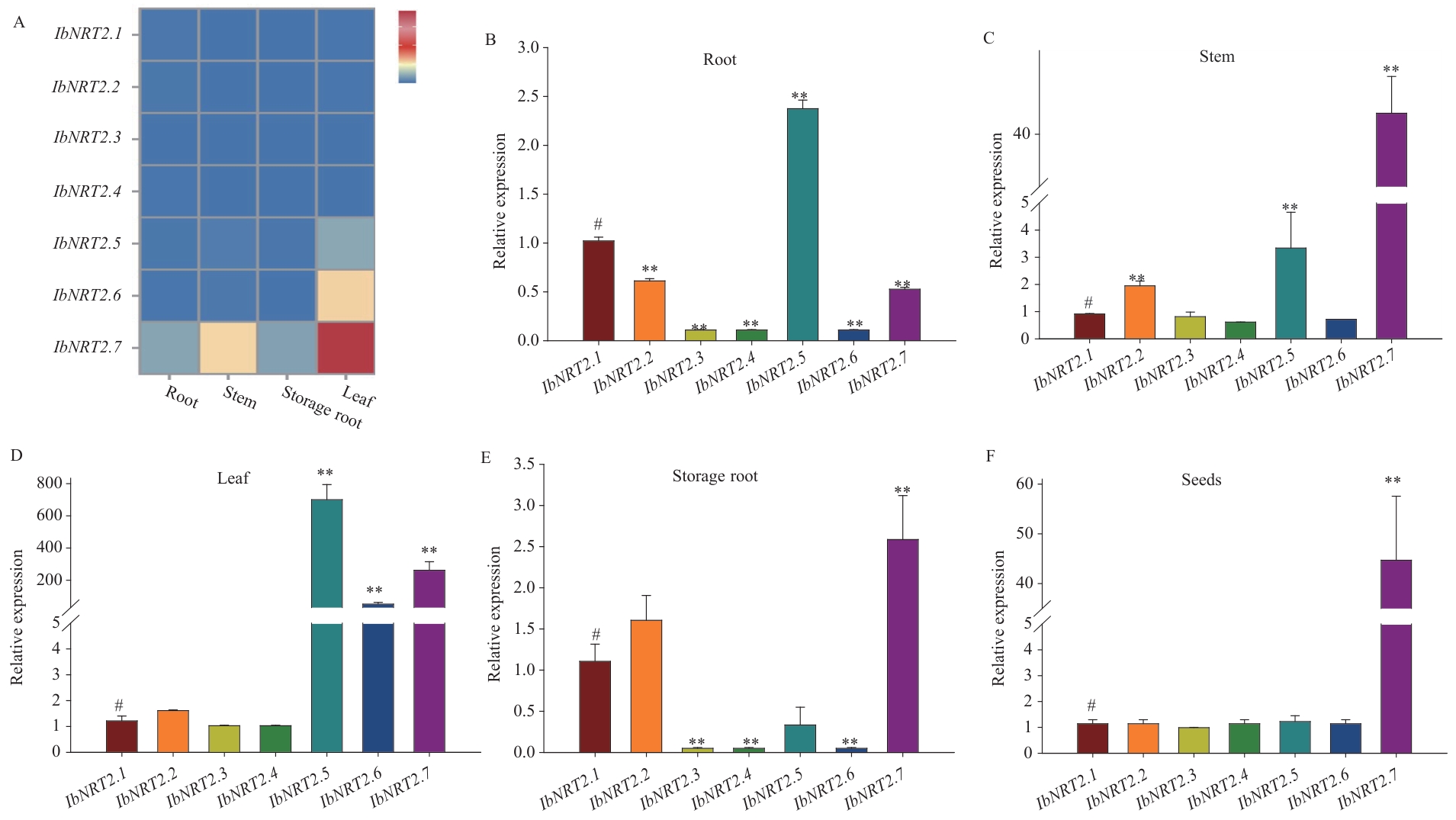
Fig. 6 Expression analysis of IbNRT2 family members in different tissuesA: Heat maps of different tissue expression via transcriptome analysis, the value indicates the FPKM value; B-F: RT-qPCR analying the relative expressions of IbNRT2s in different tissues, # indicates control, ** P<0.01

Fig. 7 Expression analysis of IbNRT2 family members under salt stressA: Expression induced by salt stress in the shoot. B: Expression induced by salt stress in the root. The concentration of NaCl in the treatment is 100 mmol/L

Fig. 8 Low nitrogen phenotype screening of different sweet potato materials (A), biomass statistical analysis (B-D) and physiological index detection (E-G)
| [1] | Frink CR, Waggoner PE, Ausubel JH. Nitrogen fertilizer: retrospect and prospect [J]. Proc Natl Acad Sci USA, 1999, 96(4): 1175-1180. |
| [2] | 储成才, 王毅, 王二涛. 植物氮磷钾养分高效利用研究现状与展望 [J]. 中国科学:生命科学, 2021, 51 (10): 1415-1423. |
| Chu CC, Wang Y, Wang ET. Improving the utilization efficiency of nitrogen, phosphorus and potassium: current situation and future perspectives [J]. Sci Sin (Vitae), 2021, 51(10): 1415-1423. | |
| [3] | 段永康, 杨海燕, 吴文龙, 等. 植物氮素吸收、转运和同化的分子机制 [J]. 福建农业学报, 2022, 37(4): 547-554. |
| Duan YK, Yang HY, Wu WL, et al. Molecular mechanisms of nitrogen absorption, transport, and assimilation in plants [J]. Fujian J Agric Sci, 2022, 37(4): 547-554. | |
| [4] | Du RJ, Wu ZX, Yu ZX, et al. Genome-wide characterization of high-affinity nitrate transporter 2 (NRT2) gene family in Brassica napus [J]. Int J Mol Sci, 2022, 23(9): 4965. |
| [5] | Crawford NM, Forde BG. Molecular and developmental biology of inorganic nitrogen nutrition [J]. Arabidopsis Book, 2002, 1: e0011. |
| [6] | You HG, Liu YM, Minh TN, et al. Genome-wide identification and expression analyses of nitrate transporter family genes in wild soybean (Glycine soja) [J]. J Appl Genet, 2020, 61(4): 489-501. |
| [7] | O'Brien JA, Vega A, Bouguyon E, et al. Nitrate transport, sensing, and responses in plants [J]. Mol Plant, 2016, 9(6): 837-856. |
| [8] | Yan NE. Structural biology of the major facilitator superfamily transporters [J]. Annu Rev Biophys, 2015, 44: 257-283. |
| [9] | Wang YY, Hsu PK, Tsay YF. Uptake, allocation and signaling of nitrate [J]. Trends Plant Sci, 2012, 17(8): 458-467. |
| [10] | Zou X, Liu MY, Wu WH, et al. Phosphorylation at Ser28 stabilizes the Arabidopsis nitrate transporter NRT2.1 in response to nitrate limitation [J]. J Integr Plant Biol, 2020, 62(6): 865-876. |
| [11] | Liu KH, Huang CY, Tsay YF. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake [J]. Plant Cell, 1999, 11(5): 865-874. |
| [12] | Quesada A, Krapp A, Trueman LJ, et al. PCR-identification of a Nicotiana plumbaginifolia cDNA homologous to the high-affinity nitrate transporters of the crnA family [J]. Plant Mol Biol, 1997, 34(2): 265-274. |
| [13] | Trueman LJ, Richardson A, Forde BG. Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans [J]. Gene, 1996, 175(1-2): 223-231. |
| [14] | Orsel M, Krapp A, Daniel-Vedele F. Analysis of the NRT2 nitrate transporter family in Arabidopsis. structure and gene expression [J]. Plant Physiol, 2002, 129(2): 886-896. |
| [15] | Xu N, Cheng L, Kong Y, et al. Functional analyses of the NRT2 family of nitrate transporters in Arabidopsis [J]. Front Plant Sci, 2024, 15: 1351998. |
| [16] | Deng QY, Luo JT, Zheng JM, et al. Genome-wide systematic characterization of the NRT2 gene family and its expression profile in wheat (Triticum aestivum L.) during plant growth and in response to nitrate deficiency [J]. BMC Plant Biol, 2023, 23(1): 353. |
| [17] | Araki R, Hasegawa H. Expression of rice (Oryza sativa L.) genes involved in high-affinity nitrate transport during the period of nitrate induction [J]. Breed Sci, 2006, 56(3): 295-302. |
| [18] | Feng HM, Yan M, Fan XR, et al. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status [J]. J Exp Bot, 2011, 62(7): 2319-2332. |
| [19] | Jia LH, Hu DS, Wang JB, et al. Genome-wide identification and functional analysis of nitrate transporter genes (NPF, NRT2 and NRT3) in maize [J]. Int J Mol Sci, 2023, 24(16): 12941. |
| [20] | Li WB, Wang Y, Okamoto M, et al. Dissection of the AtNRT2.1: AtNRT2.2 inducible high-affinity nitrate transporter gene cluster [J]. Plant Physiol, 2007, 143(1): 425-433. |
| [21] | Kiba T, Feria-Bourrellier AB, Lafouge F, et al. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants [J]. Plant Cell, 2012, 24(1): 245-258. |
| [22] | Lezhneva L, Kiba T, Feria-Bourrellier AB, et al. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants [J]. Plant J, 2014, 80(2): 230-241. |
| [23] | Chopin F, Orsel M, Dorbe MF, et al. The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds [J]. Plant Cell, 2007, 19(5): 1590-1602. |
| [24] | Yan M, Fan XR, Feng HM, et al. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges [J]. Plant Cell Environ, 2011, 34(8): 1360-1372. |
| [25] | Kwak SS. Biotechnology of the sweetpotato: ensuring global food and nutrition security in the face of climate change [J]. Plant Cell Rep, 2019, 38(11): 1361-1363. |
| [26] | 段文学, 张海燕, 解备涛, 等. 甘薯氮素营养研究进展 [J]. 西北农业学报, 2015, 24(12): 14-23. |
| Duan WX, Zhang HY, Xie BT, et al. Research advances of nitrogen nutrition in sweet potato [J]. Acta Agric Boreali Occidentalis Sin, 2015, 24(12): 14-23. | |
| [27] | Iqbal A, Dong Q, Alamzeb M, et al. Untangling the molecular mechanisms and functions of nitrate to improve nitrogen use efficiency [J]. J Sci Food Agric, 2020, 100(3): 904-914. |
| [28] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [29] | Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11 [J]. Mol Biol Evol, 2021, 38(7): 3022-3027. |
| [30] | Wang YP, Tang HB, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity [J]. Nucleic Acids Res, 2012, 40(7): e49. |
| [31] | Liu QC. Improvement for agronomically important traits by gene engineering in sweetpotato [J]. Breed Sci, 2017, 67(1): 15-26. |
| [32] | Guo K, Bian XF, Jia ZD, et al. Effects of nitrogen level on structural and functional properties of starches from different colored-fleshed root tubers of sweet potato [J]. Int J Biol Macromol, 2020, 164: 3235-3242. |
| [33] | Yang J, Moeinzadeh MH, Kuhl H, et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history [J]. Nat Plants, 2017, 3(9): 696-703. |
| [34] | Yan MX, Li M, Wang YZ, et al. Haplotype-based phylogenetic analysis and population genomics uncover the origin and domestication of sweetpotato [J]. Mol Plant, 2024, 17(2): 277-296. |
| [35] | Shi XL, Cui F, Han XY, et al. Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen-use efficiency in the wheat cultivar Kenong 9204 [J]. Mol Plant, 2022, 15(9): 1440-1456. |
| [36] | Chen XB, Yao QF, Gao XH, et al. Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition [J]. Curr Biol, 2016, 26(5): 640-646. |
| [37] | van Gelderen K, Kang C, Li PJ, et al. Regulation of lateral root development by shoot-sensed far-red light via HY5 is nitrate-dependent and involves the NRT2.1 nitrate transporter [J]. Front Plant Sci, 2021, 12: 660870. |
| [38] | Porco S, Yu S, Liang T, et al. The clock-associated LUX ARRHYTHMO regulates high-affinity nitrate transport in Arabidopsis roots [J]. Plant J, 2024, 120(5): 1786-1797. |
| [1] | LI Cai-xia, LI Yi, MU Hong-xiu, LIN Jun-xuan, BAI Long-qiang, SUN Mei-hua, MIAO Yan-xiu. Identification and Bioinformatics Analysis of the bHLH Transcription Factor Family in Cucurbita moschata Duch. [J]. Biotechnology Bulletin, 2025, 41(1): 186-197. |
| [2] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [3] | TAO Na, LI Mao-xing, GUO Hua-chun. Optimization of Sweet Potato Genetic Transformation System Mediated by Agrobacterium rhizogenes [J]. Biotechnology Bulletin, 2023, 39(10): 175-183. |
| [4] | GAO Bo, MA Juan, LI Xiu-hua, LI Jiao-sheng, WANG Rong-yan, CHEN Shu-long. Cloning and Functional Analysis of Dd-mel-26 Gene from Ditylenchus destructor [J]. Biotechnology Bulletin, 2021, 37(7): 107-117. |
| [5] | Zhang Peng. Trends and Prospect of Basic Research on Root and Tuber Crops #br#in China [J]. Biotechnology Bulletin, 2015, 31(4): 65-71. |
| [6] | Liu Qian, Qi Jiying, Han Jing, Wan Yue, Zheng Shibin. Optimization of Refluxing Extraction Technology of Anthocyanins from Purple Sweet Potato by Response Surface Method [J]. Biotechnology Bulletin, 2014, 30(12): 97-104. |
| [7] | Liang Xuelian,Xie Zhenwen. Research Progress and Prospect on the Genetic Mapping Strategy of Sweet Potato [J]. Biotechnology Bulletin, 2014, 30(11): 1-6. |
| [8] | Li Zhiliang, Wu Zhongyi, Wang Yuwen, Xing Haochun, Ye Jia, Zhang Xiuhai, Huang Conglin. Advancement of the Research on Transgenic Sweet Potato (Ipomoea batatas Lam.) [J]. Biotechnology Bulletin, 2013, 29(9): 1-6. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||