








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (1): 186-197.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0606
Previous Articles Next Articles
LI Cai-xia1( ), LI Yi1, MU Hong-xiu1, LIN Jun-xuan1, BAI Long-qiang1,2, SUN Mei-hua1,2, MIAO Yan-xiu1,2(
), LI Yi1, MU Hong-xiu1, LIN Jun-xuan1, BAI Long-qiang1,2, SUN Mei-hua1,2, MIAO Yan-xiu1,2( )
)
Received:2024-06-25
Online:2025-01-26
Published:2025-01-22
Contact:
MIAO Yan-xiu
E-mail:lcx6283@163.com;miaoyanxiu@163.com
LI Cai-xia, LI Yi, MU Hong-xiu, LIN Jun-xuan, BAI Long-qiang, SUN Mei-hua, MIAO Yan-xiu. Identification and Bioinformatics Analysis of the bHLH Transcription Factor Family in Cucurbita moschata Duch.[J]. Biotechnology Bulletin, 2025, 41(1): 186-197.
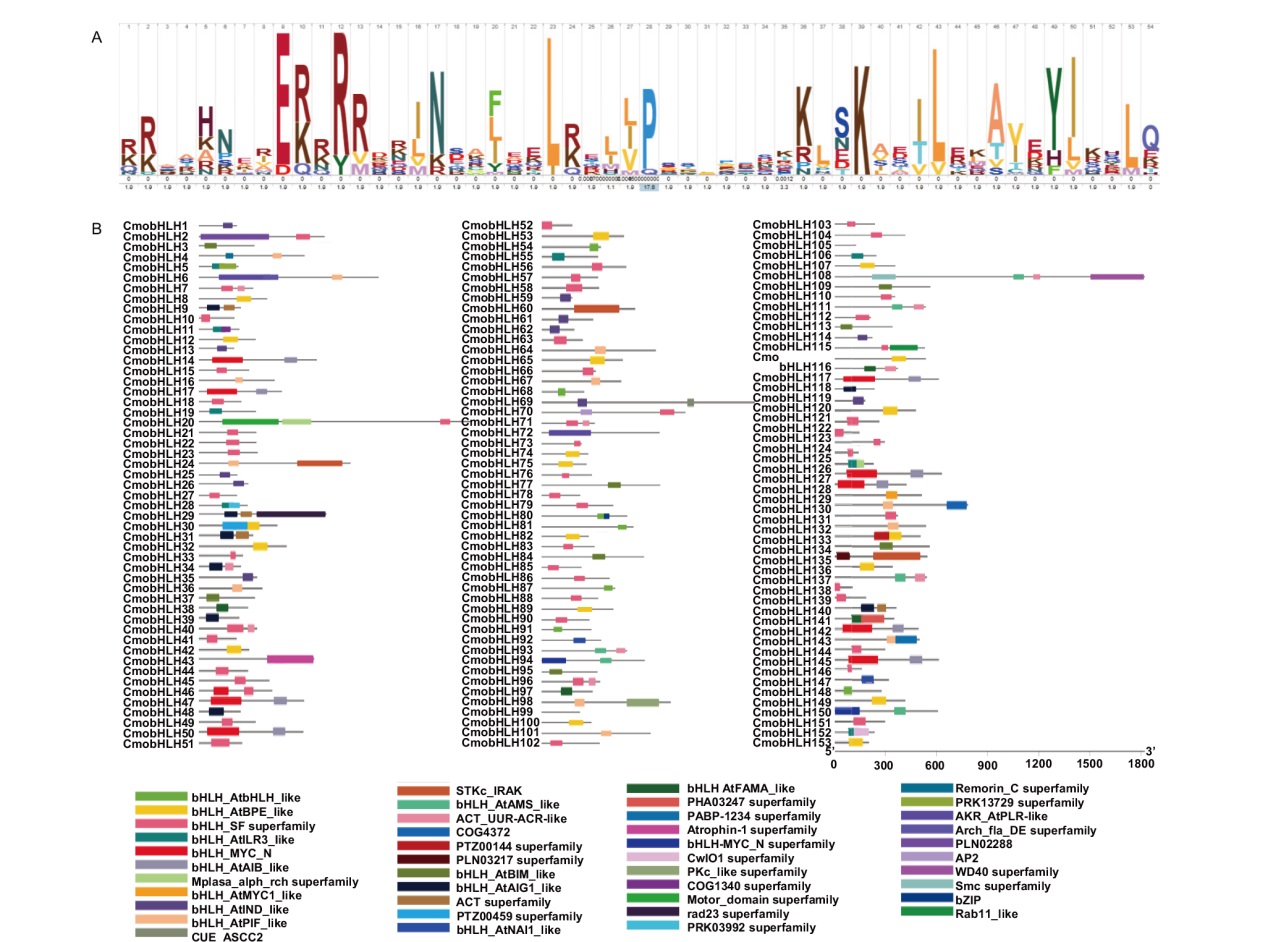
Fig. 2 HLH conservative domain and distribution of bHLH family in C. moschata Duch. A: HLH conserved domain, where the height of amino acid letters at different sites indicates the level of domain conservation. B: Distribution of 43 other conserved domains of bHLH family in C. moschata Duch.

Fig. 3 Phylogenetic and structural analysis of the bHLH family in C. moschata Duch. A: Phylogenetic tree. B: Gene motif analysis. C: Intron and exon distribution. Introns are indicated by black lines, exons are indicated by green boxes. The scale bar is shown at the bottom
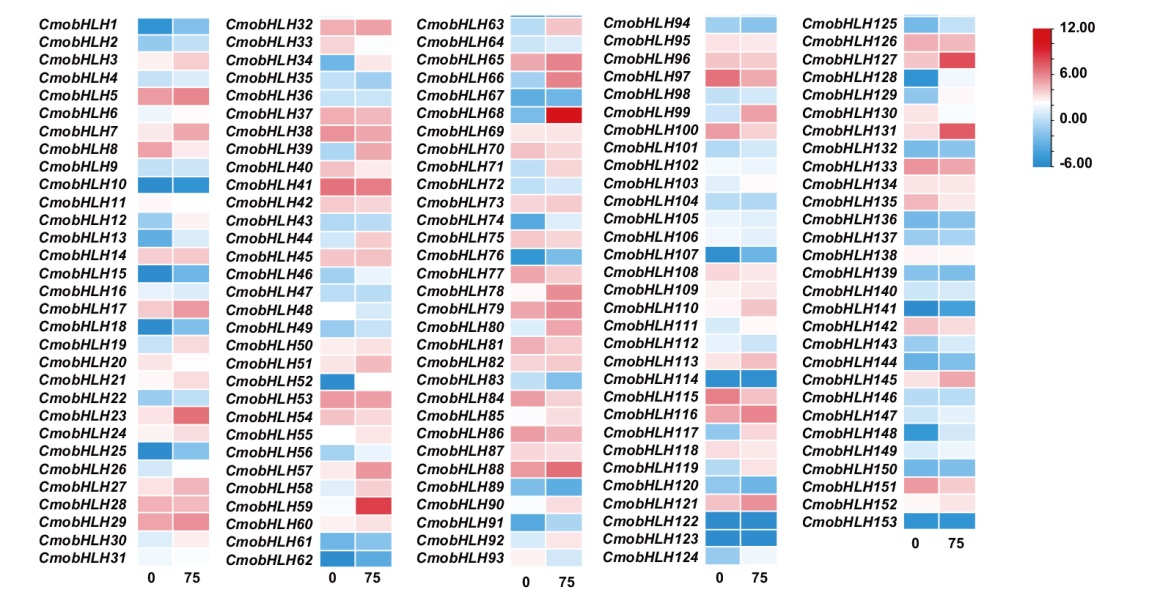
Fig. 6 Expression analysis of bHLH gene family in response to salt stress in C. moschata Duch. “0” indicates 0 mmol/L NaCl; “75” indicates 75 mmol/L NaCl
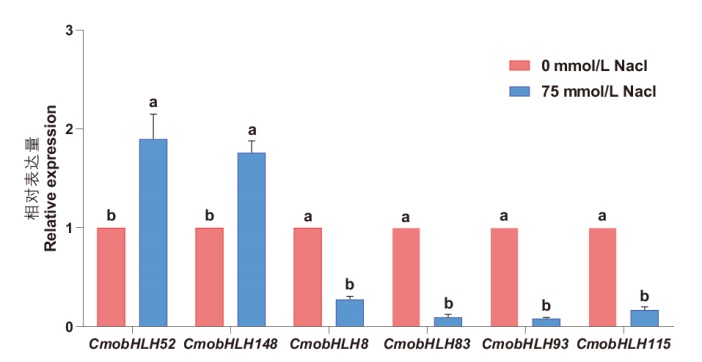
Fig. 7 RT-qPCR validation of bHLH gene family in response to salt stress in C. moschata Duch. Different lower letters indicate significant difference at P<0.05 level
| [1] |
Robinson KA, Koepke JI, Kharodawala M, et al. A network of yeast basic helix-loop-helix interactions[J]. Nucleic Acids Res, 2000, 28(22): 4460-4466.
doi: 10.1093/nar/28.22.4460 pmid: 11071933 |
| [2] |
Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins[J]. Cell, 1989, 56(5): 777-783.
doi: 10.1016/0092-8674(89)90682-x pmid: 2493990 |
| [3] |
Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis[J]. Genome Res, 2001, 11(5): 754-770.
doi: 10.1101/gr.177001 pmid: 11337472 |
| [4] |
Jones S. An overview of the basic helix-loop-helix proteins[J]. Genome Biol, 2004, 5(6): 226.
doi: 10.1186/gb-2004-5-6-226 pmid: 15186484 |
| [5] |
Heim MA, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity[J]. Mol Biol Evol, 2003, 20(5): 735-747.
doi: 10.1093/molbev/msg088 pmid: 12679534 |
| [6] | Li XX, Duan XP, Jiang HX, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis[J]. Plant Physiol, 2006, 141(4): 1167-1184. |
| [7] | 唐文武, 吴秀兰, 钟佩桥. 白菜bHLH转录因子家族的全基因鉴定及表达特征分析[J]. 江西农业学报, 2020, 32(6): 1-5. |
| Tang WW, Wu XL, Zhong PQ. Genome-wide identification and expression analysis of bHLH transcription factor family in B. rapa[J]. Acta Agric Jiangxi, 2020, 32(6): 1-5. | |
| [8] | 郭鹏宇. 番茄bHLH转录因子SlPRE3的克隆与功能研究[D]. 重庆: 重庆大学, 2021. |
| Guo PY. Cloning and functional study of tomato bHLH transcription factor SlPRE3[D]. Chongqing: Chongqing University, 2021. | |
| [9] | 薛宝平. 辣椒bHLH基因家族的鉴定、表达分析及CabHLH94在应答青枯菌侵染中的作用[D]. 延安: 延安大学, 2019. |
| Xue BP. Identification and expression analysis of bHLH gene family in pepper and the role of CabHLH94 in responding to Ralstonia solanacearum infection[D]. Yan'an: Yan'an University, 2019. | |
| [10] | 张鲲, 史后蕊, 马婧, 等. 花生bHLH基因家族的鉴定与表达分析[J/OL]. 分子植物育种, 2022. http://kns.cnki.net/kcms/detail/46.1068.S.20221121.0836.002.html |
| Zhang K, Shi HR, Ma J, et al. Identification and expression analysis of bHLH gene family in peanut[J/OL]. Mol Plant Breed, 2022. http://kns.cnki.net/kcms/detail/46.1068.S.20221121.0836.002.html | |
| [11] | 何开平, 吴楚. bHLH转录因子对植物形态发生的影响[J]. 安徽农业科学, 2010, 38(35): 19957-19959. |
| He KP, Wu C. The effects of bHLH transcription factors on plant morphogenesis[J]. J Anhui Agric Sci, 2010, 38(35): 19957-19959. | |
| [12] | 唐琪琦, 李金帅, 邱帅, 等. 绣球花型全基因组关联分析[J]. 植物遗传资源学报, 2023, 24(4): 1174-1185. |
| Tang QQ, Li JS, Qiu S, et al. Genome-wide association study of Hydrangea macrophylla inflorescence type[J]. J Plant Genet Resour, 2023, 24(4): 1174-1185. | |
| [13] | Xiang LL, Liu XF, Shi YN, et al. Comparative transcriptome analysis revealed two alternative splicing bHLHs account for flower color alteration in Chrysanthemum[J]. Int J Mol Sci, 2021, 22(23): 12769. |
| [14] |
Zhang F, Gonzalez A, Zhao MZ, et al. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis[J]. Development, 2003, 130(20): 4859-4869.
doi: 10.1242/dev.00681 pmid: 12917293 |
| [15] | Liu WW, Tai HH, Li SS, et al. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism[J]. New Phytol, 2014, 201(4): 1192-1204. |
| [16] | Wang PF, Su L, Gao HH, et al. Genome-wide characterization of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis[J]. Front Plant Sci, 2018, 9: 64. |
| [17] | Shen TJ, Wen XP, Wen Z, et al. Genome-wide identification and expression analysis of bHLH transcription factor family in response to cold stress in sweet cherry(Prunus avium L.)[J]. Sci Hortic, 2021, 279: 109905. |
| [18] | Dong Y, Wang CP, Han X, et al. A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis[J]. Biochem Biophys Res Commun, 2014, 450(1): 453-458. |
| [19] | 徐颖超, 张思程, 薛舒丹, 等. 南瓜叶黄素基因紧密连锁的InDel分子标记开发及应用[J]. 江苏农业学报, 2024, 40(2): 348-358. |
| Xu YC, Zhang SC, Xue SD, et al. Development and application of closely linked InDel molecular markers of lutein gene in Cucurbita moschata Duch[J]. Jiangsu J Agric Sci, 2024, 40(2): 348-358. | |
| [20] | 杨建坤, 殷缘, 车易达, 等. 胁迫条件下山新杨PdPapMYC2基因的组织表达模式[J]. 西北农林科技大学学报: 自然科学版, 2023, 51(6): 18-27. |
| Yang JK, Yin Y, Che YD, et al. Tissue-specific expression profiles of PdPapMYC2 in Shanxin poplar under various stresses[J]. J Northwest A F Univ Nat Sci Ed, 2023, 51(6): 18-27. | |
| [21] |
隋心意, 赵小刚, 毛欣, 等. 生菜bHLH转录因子家族的鉴定与生物信息学分析[J]. 中国农学通报, 2023, 39(3): 104-110.
doi: 10.11924/j.issn.1000-6850.casb2022-0214 |
| Sui XY, Zhao XG, Mao X, et al. Identification and bioinformatics analysis of bHLH transcription factor family in Lactuca sativaL[J]. Chin Agric Sci Bull, 2023, 39(3): 104-110. | |
| [22] | Eailey TL, Elkan C. Proceedings of the second international conference on intelligent systems for molecular biology[C]. Menlo Park, California: AAAI Press, 1994: 28-36. |
| [23] | 李君霞, 樊永强, 代书桃, 等. bHLH转录因子在植物抗非生物胁迫基因工程中的应用进展[J]. 江苏农业科学, 2022, 50(12): 1-9. |
| Li JX, Fan YQ, Dai ST, et al. Application progress of bHLH transcription factor in plant abiotic stress resistance genetic engineering[J]. Jiangsu Agric Sci, 2022, 50(12): 1-9. | |
| [24] | 高珂. 基于与BcAS1启动子互作及表达同步性筛选调控柴胡皂苷合成转录因子[D]. 北京: 北京协和医学院, 2016. |
| Gao K. Screening transcription factors regulating saikosaponin synthesis based on interaction with BcAS1 promoter and synchronization of expression[D]. Beijing: Peking Union Medical College, 2016. | |
| [25] |
Bailey PC, Martin C, Toledo-Ortiz G, et al. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana[J]. Plant Cell, 2003, 15(11): 2497-2502.
pmid: 14600211 |
| [26] | Tan C, Qiao HL, Ma M, et al. Genome-wide identification and characterization of melon bHLH transcription factors in regulation of fruit development[J]. Plants, 2021, 10(12): 2721. |
| [27] | 丁冬, 李嘉欣, 魏玉磊, 等. 拟南芥和玉米中膜结合bHLH转录因子的鉴定与分析[J]. 植物生理学报, 2020, 56(4): 700-710. |
| Ding D, Li JX, Wei YL, et al. Identification and analysis of membrane-bound bHLH transcription factors in Arabidopsis thaliana and maize[J]. Plant Physiol J, 2020, 56(4): 700-710. | |
| [28] | 李晓星, 蒋海雄, 黄青云, 等. 水稻bHLH基因家族成员表达模式分析[J]. 厦门大学学报: 自然科学版, 2006, S1: 73-77. |
| Li XX, Jiang HX, Huang QY, et al. Expression pattern analysis of rice bHLH gene family members[J]. J Xiamen Univ Nat Sci, 2006, S1: 73-77. | |
| [29] | 王艳敏, 白卉, 曹焱. bHLH转录因子研究进展及其在植物抗逆中的应用[J]. 安徽农业科学, 2015, 43(21): 34-35, 50. |
| Wang YM, Bai H, Cao Y. Research progress of bHLH transcription factor and application in plant abiotic stress tolerance[J]. J Anhui Agric Sci, 2015, 43(21): 34-35, 50. | |
| [30] |
范子培, 李龙, 史雨刚, 等. 小麦TabHLH112-2B基因克隆及每穗小穗数相关功能标记开发[J]. 作物学报, 2024, 50(2): 403-413.
doi: 10.3724/SP.J.1006.2024.31016 |
| Fan ZP, Li L, Shi YG, et al. Cloning of TabHLH112-2B gene and development of its functional marker associated with the number of spikelet per spike in wheat[J]. Acta Agron Sin, 2024, 50(2): 403-413. | |
| [31] | 刘选明, 彭玉冲, 杨远柱, 等. 水稻 bHLH转录因子 Os11g39000的功能研究[J]. 湖南大学学报: 自然科学版, 2016, 43(6): 109-116. |
| Liu XM, Peng YC, Yang YZ, et al. Functional analysis of a bHLH transcription factor Os11g39000 in rice[J]. J Hunan Univ Nat Sci, 2016, 43(6): 109-116. | |
| [32] | Zhou J, Li F, Wang JL, et al. Basic helix-loop-helix transcription factor from wild rice(OrbHLH2)improves tolerance to salt- and osmotic stress in Arabidopsis[J]. J Plant Physiol, 2009, 166(12): 1296-1306. |
| [33] | 吕宝莲, 杨宇昕, 崔立操, 等. 小麦bHLH家族转录因子的鉴定及其在盐胁迫条件下的表达分析[J]. 作物杂志, 2024(1): 65-72. |
| Lü BL, Yang YX, Cui LC, et al. Identification of bHLH family transcription factors of wheat and expression analysis under salt stress[J]. Crops, 2024(1): 65-72. | |
| [34] | 朱涛, 李芳菲, 杨海涵, 等. 山药bHLH基因家族鉴定及表达分析[J]. 信阳师范学院学报: 自然科学版, 2022, 35(3): 393-399. |
| Zhu T, Li FF, Yang HH, et al. Identification and expression ananlysis of bHLH gene family in Chinese yam(Dioscoreae Rhizoma)[J]. J Xinyang Norm Univ Nat Sci Ed, 2022, 35(3): 393-399. |
| [1] | LI Yu-qing, WU Nan, LUO Jian-rang. Cloning and Functional Analysis of bHLH Gene Related to Anthocyanin Synthesis in Paeonia qiui [J]. Biotechnology Bulletin, 2024, 40(8): 174-185. |
| [2] | ZHOU Lin, HUANG Shun-man, SU Wen-kun, YAO Xiang, QU Yan. Identification of the bHLH Gene Family and Selection of Genes Related to Color Formation in Camellia reticulata [J]. Biotechnology Bulletin, 2024, 40(8): 142-151. |
| [3] | HU Yong-bo, LEI Yu-tian, YANG Yong-sen, CHEN Xin, LIN Huang-fang, LIN Bi-ying, LIU Shuang, BI Ge, SHEN Bao-ying. Genome-wide Identification and Expression Pattern Analysis of the Bcl-2-related Anti-apoptotic Family in Cucumis sativus L. and Cucurbita moschata Duch. [J]. Biotechnology Bulletin, 2024, 40(6): 219-237. |
| [4] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [5] | LIU Bao-cai, ZHANG Wu-jun, ZHAO Yun-qing, HUANG Ying-zhen, CHEN Jing-ying. Identification and Tissue Expression of Four Transcription Factors in Pholidota chinensis [J]. Biotechnology Bulletin, 2024, 40(5): 141-152. |
| [6] | WANG Bin, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui, HE Jin-ming. Cloning of bHLH96 Gene and Its Roles in Regulating the Biosynthesis of Peppermint Terpenes [J]. Biotechnology Bulletin, 2024, 40(1): 281-293. |
| [7] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [8] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [9] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [10] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [11] | LIU Cheng-xia, SUN Zong-yan, LUO Yun-bo, ZHU Hong-liang, QU Gui-qin. Multifaceted Roles of bHLH Phosphorylation in Regulation of Plant Physiological Functions [J]. Biotechnology Bulletin, 2023, 39(3): 26-34. |
| [12] | YAN Meng-yu, WEI Xiao-wei, CAO Jing, LAN Hai-yan. Cloning of Basic Helix-loop-helix(bHLH)Transcription Factor Gene SabHLH169 in Suaeda aralocaspica and Analysis of Its Resistances to Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 328-339. |
| [13] | AN Chang, LU Lin, SHEN Meng-qian, CHEN Sheng-zhen, YE Kang-zhuo, QIN Yuan, ZHENG Ping. Research Progress of bHLH Gene Family in Plants and Its Application Prospects in Medical Plants [J]. Biotechnology Bulletin, 2023, 39(10): 1-16. |
| [14] | FENG Jian-ying, LI Li-qin, LU Li-ming. Genome-wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Solanum tuberosum [J]. Biotechnology Bulletin, 2022, 38(2): 21-33. |
| [15] | SHAN Cao-mei, YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang. Cloning,and Functional Analysis of Gene OsRAI1 Resistant to Hirschmanniella mucronate in Rice [J]. Biotechnology Bulletin, 2021, 37(7): 146-155. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||