








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 195-206.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0231
XU Xiao-ping1( ), YANG Cheng-long1, HE Xing1,2, GUO Wen-jie1, WU Jian3, FANG Shao-zhong1(
), YANG Cheng-long1, HE Xing1,2, GUO Wen-jie1, WU Jian3, FANG Shao-zhong1( )
)
Received:2025-03-05
Online:2025-09-26
Published:2025-09-24
Contact:
FANG Shao-zhong
E-mail:byxxp310107@163.com;fangshaozhong@faas.cn
XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium[J]. Biotechnology Bulletin, 2025, 41(9): 195-206.
引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) | 退火温度 Annealing temperature (℃) | 延伸时间Extended time | 用途 Application |
|---|---|---|---|---|
| LoAPS1-F | ATGGCCACCATGGCTGCCCTC | 61 | 1 min 45 s | LoAPS1 克隆 LoAPS1 cloning |
| LoAPS1-R | TCAAGCTGGCACAGCCTCACGC | |||
| LoAPS1-1 302-F | ggcatggtagatctgACTAGTATGGCCACCATGGCTGCC | 60 | 1 min 45 s | LoAPS1亚细胞定位 LoAPS1 subcellular localization |
| LoAPS1-1 302-R | aagttcttctcctttACTAGTAGCTGGCACAGCCTCACGCA | |||
| TRV2-LoAPS1-F | tgagtaaggttaccgGAATTCCCCGATCTTATTGCTTCACC | 57 | 35 s | LoAPS1沉默片段扩增 LoAPS1 silent segment amplification |
| TRV2-LoAPS1-R | gggacatgcccgggcCTCGAGAGGCAACACTGTCATAGTACTC | |||
| TRV2-Coat protein-F | CTAACAGTGCTCTTGGTGTGATT | 55 | 25 s | TRV2病毒检测 TRV2 virus detection |
| TRV2-Coat protein-R | CAACTCCATGTTCTCTAACGAAGT | |||
| LoAPS1-qF | ATGGGTCCTTCGTCAACATG | 60 | 30 s | LoAPS1 qPCR扩增 LoAPS1 qPCR amplification |
| LoAPS1-qR | CAGCATTGGTGATAGCTTCCT | |||
| Loβ-Actin-qF | CGGTGTCTGGATTGGAGGGTCA | 60 | 30 s | qPCR扩增内参基因 Amplification of internal reference genes by qPCR |
| Loβ-Actin-qR | TTCGCTTTAGGACTTCGGGT |
Table 1 Primer sequences of gene cloning, subcellular localization and gene silence
引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) | 退火温度 Annealing temperature (℃) | 延伸时间Extended time | 用途 Application |
|---|---|---|---|---|
| LoAPS1-F | ATGGCCACCATGGCTGCCCTC | 61 | 1 min 45 s | LoAPS1 克隆 LoAPS1 cloning |
| LoAPS1-R | TCAAGCTGGCACAGCCTCACGC | |||
| LoAPS1-1 302-F | ggcatggtagatctgACTAGTATGGCCACCATGGCTGCC | 60 | 1 min 45 s | LoAPS1亚细胞定位 LoAPS1 subcellular localization |
| LoAPS1-1 302-R | aagttcttctcctttACTAGTAGCTGGCACAGCCTCACGCA | |||
| TRV2-LoAPS1-F | tgagtaaggttaccgGAATTCCCCGATCTTATTGCTTCACC | 57 | 35 s | LoAPS1沉默片段扩增 LoAPS1 silent segment amplification |
| TRV2-LoAPS1-R | gggacatgcccgggcCTCGAGAGGCAACACTGTCATAGTACTC | |||
| TRV2-Coat protein-F | CTAACAGTGCTCTTGGTGTGATT | 55 | 25 s | TRV2病毒检测 TRV2 virus detection |
| TRV2-Coat protein-R | CAACTCCATGTTCTCTAACGAAGT | |||
| LoAPS1-qF | ATGGGTCCTTCGTCAACATG | 60 | 30 s | LoAPS1 qPCR扩增 LoAPS1 qPCR amplification |
| LoAPS1-qR | CAGCATTGGTGATAGCTTCCT | |||
| Loβ-Actin-qF | CGGTGTCTGGATTGGAGGGTCA | 60 | 30 s | qPCR扩增内参基因 Amplification of internal reference genes by qPCR |
| Loβ-Actin-qR | TTCGCTTTAGGACTTCGGGT |
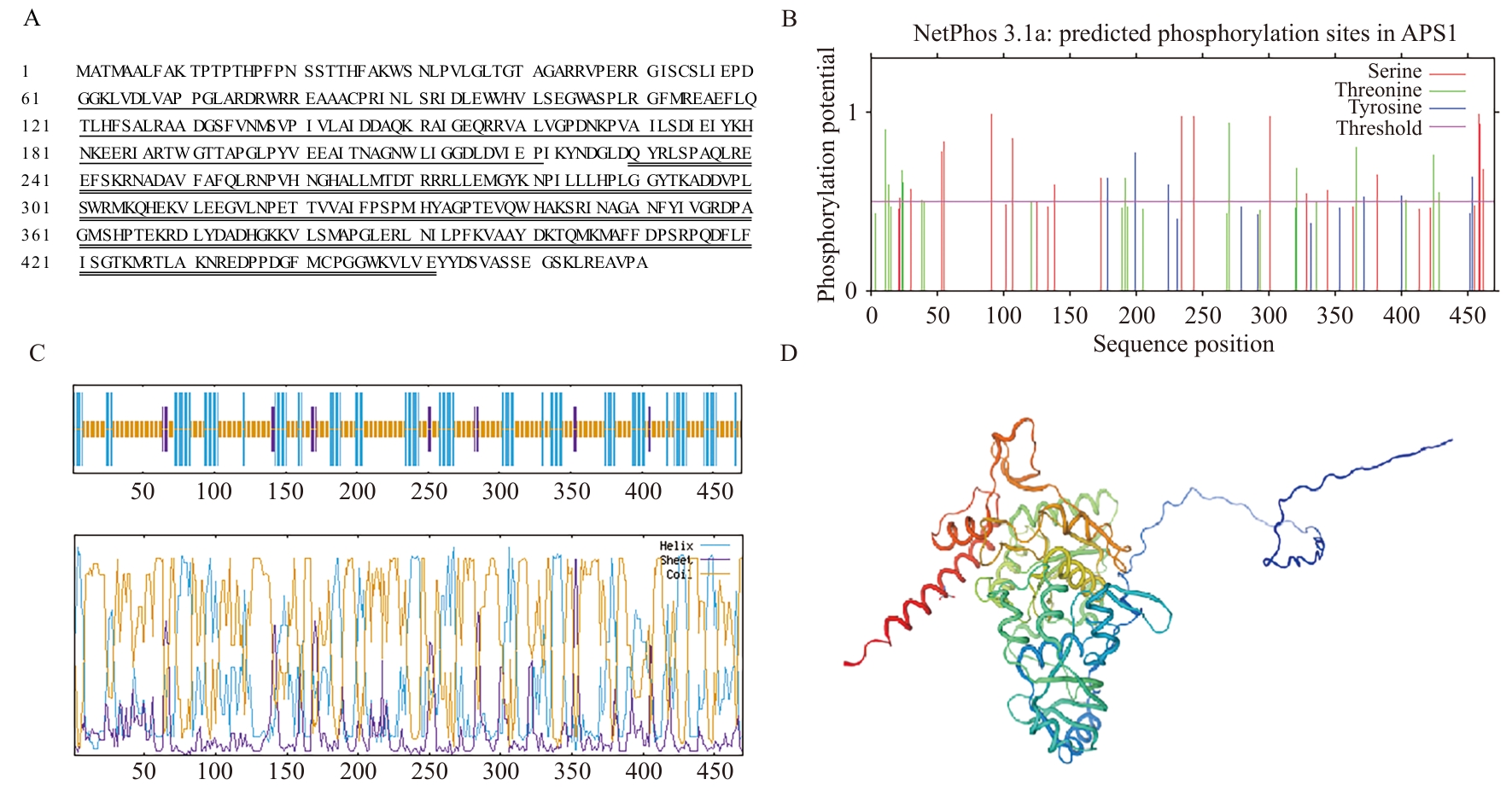
Fig. 2 Distribution of amino acids (A) in the LoAPS1 protein domains, prediction of phosphorylation sites (B), and models of secondary (C) and tertiary (D) protein structuresIn Fig. A, the single horizontal line indicates the PUA-like domain in the amino acid region from 57 to 221, while the double horizontal line indicates the ATP-sulfurylase conserved domain in the amino acid region from 229 to 452
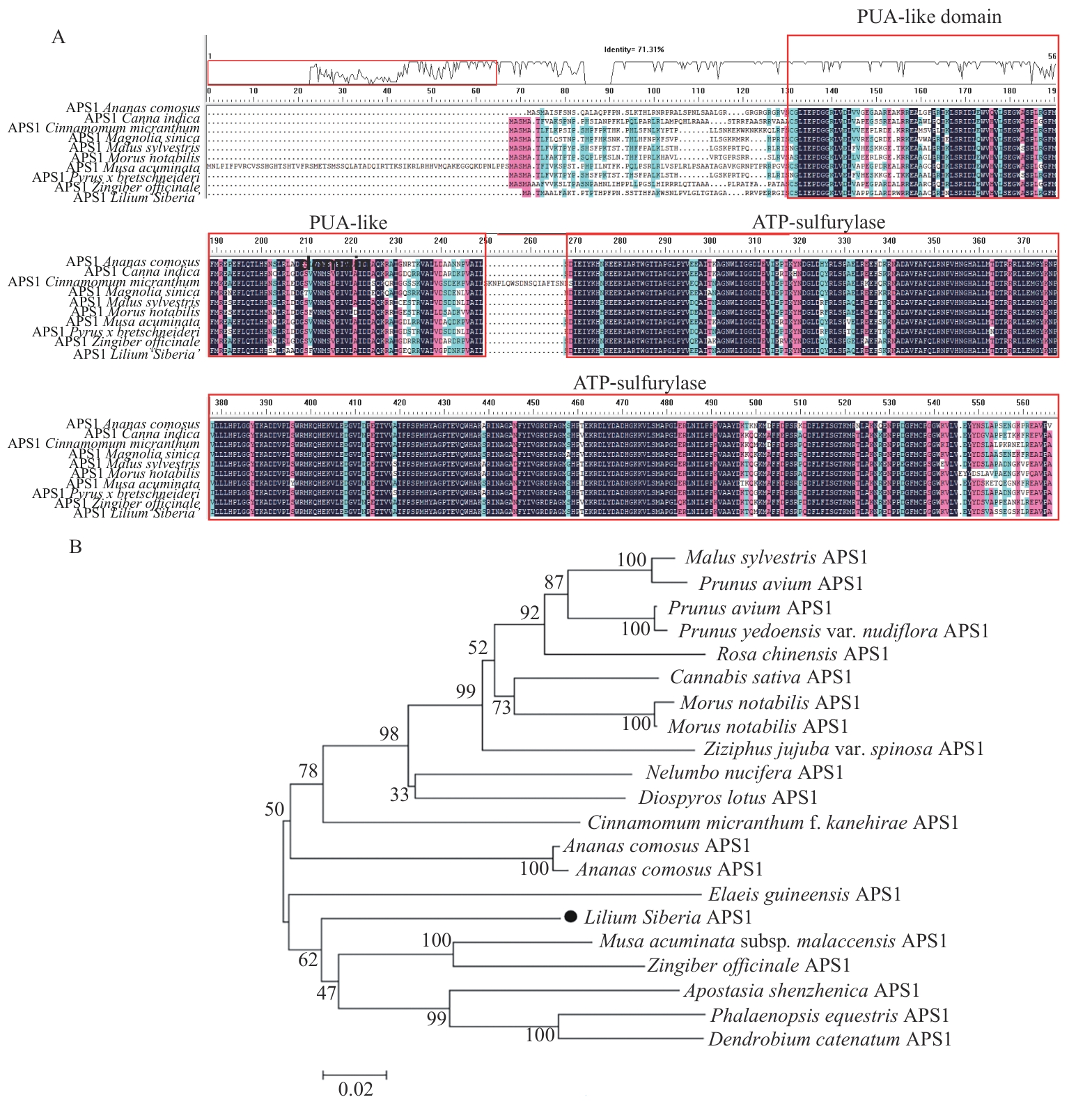
Fig. 3 Multiple sequence alignment of LoAPS1 with APS1 from other species (A) and phylogenetic tree analysis (B)The red boxes in Fig. A indicate the PUA-like domain and ATP-sulfurylase
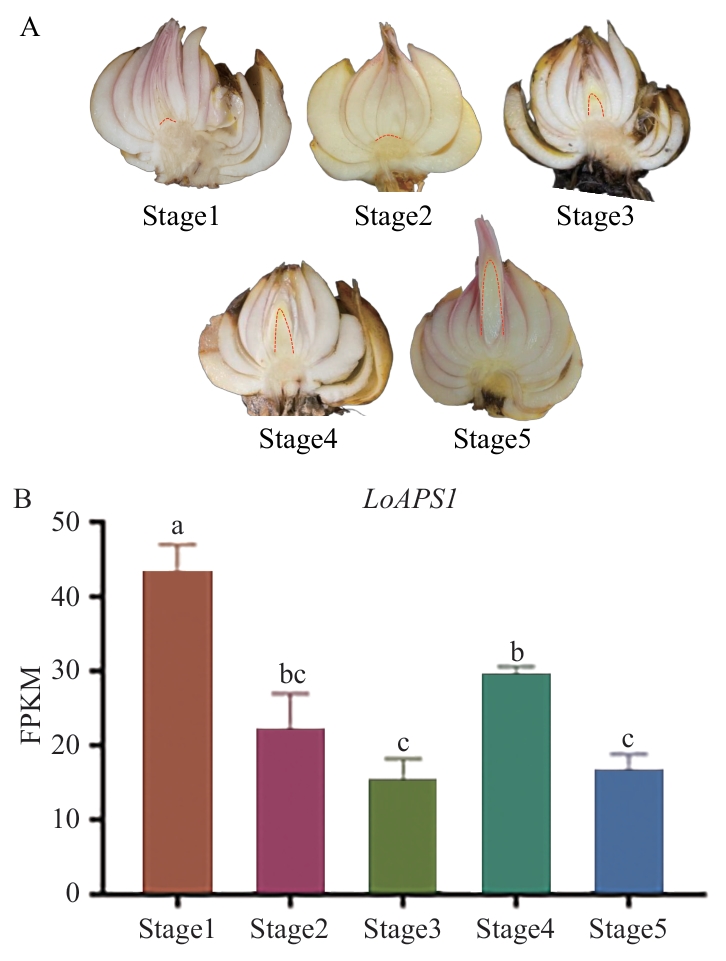
Fig. 6 Analysis of the expression pattern of LoAPS1 during different stages of lily dormancy releaseA: Longitudinal anatomical morphologies of bulbs at different stages in the dormancy release process of lily. B: Transcriptome FPKM analysis of APS1 in different stages of dormancy release of lily. Stage1: Lily bulb collection period. Stage2: Dormancy maintenance period. Stage3: Tip bud break but not reach two-thirds of the bulb. Stage4: Dormancy complete release stage. Stage5: Rapid growth stage of the tip bud. Lowercase letters indicate significant differences at the P<0.05 level

Fig. 7 Sulfur metabolism and expression patterns of related genes in the transcriptome of lily bulb during different concentrations of riboflavin treatment promoting dormancy releaseA: 0.5 mmol/L riboflavin treatment promoted significant enrichment of sulfur metabolism in KEGG metabolic pathway during dormancy release of lily bulbs. B: FPKM expression pattern analysis of sulfur metabolism-related differential genes
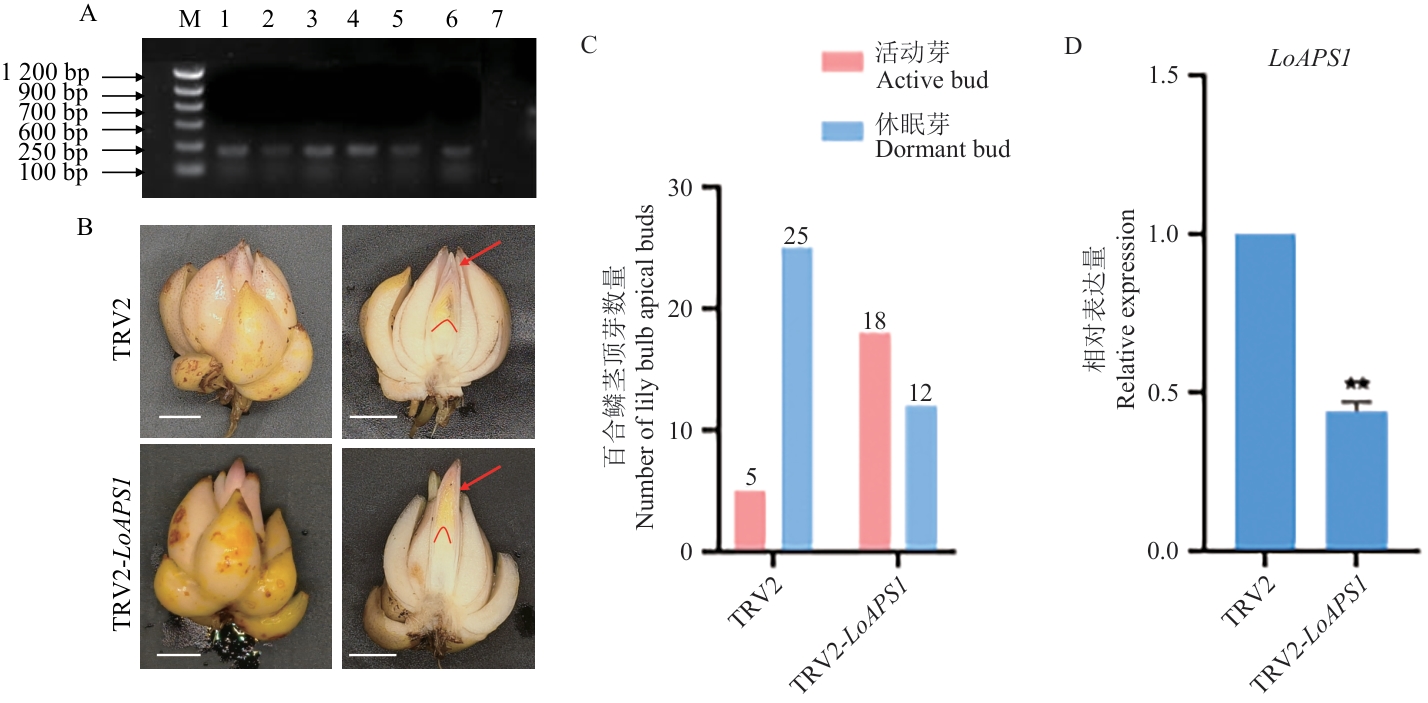
Fig. 9 Effects of LoAPS1 silencing on the lily bulb dormancy releaseA: TRV2 virus detection (M: DL 1200 DNA Maker; 1‒3: TRV2 line of bulbs; 4‒6: TRV2-LoAPS1 silenced line; 7: untreated bulb). B: Statistical analysis of the number of sprouts active bulbs after TRV2-LoAPS1 silencing. C: Phenotype diagram of TRV2 and TRV2-LoAPS1 affecting the dormancy release of lily bulbs, scale bar=1 cm, the red arrow indicates active buds. D: Relative expressions of LoAPS1 in the TRV2 and TRV2-LoAPS1 silenced lines. ** indicates significant difference at the P<0.01 level compared to TRV2
| [1] | 于险峰, 张仁军. 百合种球国产化实现新突破 [N]. 农民日报, 2022-12-20(7)[2025-04-16]. DOI:10.28603/n.cnki.nnmrb.2022.005237 . |
| Yu XF, Zhang R J. Lily ball localization achieved a new breakthrough [N] . Farmers Daily China Agricultural Network, 2022-12-20(7)[2025-04-16]. DOI: 10.28603/n.cnki.nnmrb.2022.005237 . | |
| [2] | Kopriva S, Malagoli M, Takahashi H. Sulfur nutrition: impacts on plant development, metabolism, and stress responses [J]. J Exp Bot, 2019, 70(16): 4069-4073. |
| [3] | Sun SK, Chen J, Zhao FJ. Regulatory mechanisms of sulfur metabolism affecting tolerance and accumulation of toxic trace metals and metalloids in plants [J]. J Exp Bot, 2023, 74(11): 3286-3299. |
| [4] | Takahashi H, Marsolais F, Cuypers A, et al. Sulfur metabolism: actions for plant resilience and environmental adaptation [J]. J Exp Bot, 2023, 74(11): 3271-3275. |
| [5] | Kopriva S, Rahimzadeh Karvansara P, Takahashi H. Adaptive modifications in plant sulfur metabolism over evolutionary time [J]. J Exp Bot, 2024, 75(16): 4697-4711. |
| [6] | Rouached H, Wirtz M, Alary R, et al. Differential regulation of the expression of two high-affinity sulfate transporters, SULTR1.1 and SULTR1.2, in Arabidopsis [J]. Plant Physiol, 2008, 147(2): 897-911. |
| [7] | Chen Z, Zhao PX, Miao ZQ, et al. SULTR3s function in chloroplast sulfate uptake and affect ABA biosynthesis and the stress response [J]. Plant Physiol, 2019, 180(1): 593-604. |
| [8] | Takahashi H, Kopriva S, Giordano M, et al. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes [J]. Annu Rev Plant Biol, 2011, 62: 157-184. |
| [9] | Saito K. Sulfur assimilatory metabolism. The long and smelling road [J]. Plant Physiol, 2004, 136(1): 2443-2450. |
| [10] | Logan HM, Cathala N, Grignon C, et al. Cloning of a cDNA encoded by a member of the Arabidopsis thaliana ATP sulfurylase multigene family expression studies in yeast and in relation to plant sulfur nutrition [J]. J Biol Chem, 1996, 271(21): 12227-12233. |
| [11] | Hatzfeld Y, Cathala N, Grignon C, et al. Effect of ATP sulfurylase overexpression in bright yellow 2 tobacco cells. Regulation of atp sulfurylase and SO4 2- transport activities [J]. Plant Physiol, 1998, 116(4): 1307-1313. |
| [12] | Klonus D, Höfgen R, Willmitzer L, et al. Isolation and characterization of two cDNA clones encoding ATP-sulfurylases from potato by complementation of a yeast mutant [J]. Plant J, 1994, 6(1): 105-112. |
| [13] | 张毛毛. 水稻OsmtATPS1基因的克隆及功能初步分析 [D]. 杨凌: 西北农林科技大学, 2015. |
| Zhang MM. Cloning and functional preliminary study of OsmtATPS1 in rice [D]. Yangling: Northwest A & F University, 2015. | |
| [14] | 王帮庆. 油菜ATPS基因克隆、原核表达及其活性鉴定 [D]. 杨凌: 西北农林科技大学, 2009. |
| Wang BQ. Cloning, prokaryoyic expression and activity identification of Brassica napus ATPS [D]. Yangling: Northwest A & F University, 2009. | |
| [15] | 朱磊, 杨路明, 孙守如, 等. 辣椒ATP硫酸化酶基因(CaATPS1)的克隆与逆境表达 [J]. 中国生物化学与分子生物学报, 2016, 32(9): 1040-1047. |
| Zhu L, Yang LM, Sun SR, et al. Cloning of a pepper CaATPS1 gene and its expression in response to stresses [J]. Chin J Biochem Mol Biol, 2016, 32(9): 1040-1047. | |
| [16] | Batool S, Uslu VV, Rajab H, et al. Sulfate is incorporated into cysteine to trigger ABA production and stomatal closure [J]. Plant Cell, 2018, 30(12): 2973-2987. |
| [17] | Shen J, Zhang J, Zhou MJ, et al. Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling [J]. Plant Cell, 2020, 32(4): 1000-1017. |
| [18] | Elferjani R, Pahari S, Soolanayakanahally R, et al. Drought induced metabolic shifts and water loss mechanisms in canola: role of cysteine, phenylalanine and aspartic acid [J]. Front Plant Sci, 2024, 15: 1385414. |
| [19] | Gong L, Liu HQ, Hua Y, et al. ABA-induced active stomatal closure in bulb scales of Lanzhou lily [J]. Plant Signal Behav, 2025, 20(1): e2446865. |
| [20] | Wu J, Jin YJ, Liu C, et al. GhNAC83 inhibits corm dormancy release by regulating ABA signaling and cytokinin biosynthesis in Gladiolus hybridus [J]. J Exp Bot, 2019, 70(4): 1221-1237. |
| [21] | Przybyla-Toscano J, Christ L, Keech O, et al. Iron-sulfur proteins in plant mitochondria: roles and maturation [J]. J Exp Bot, 2021, 72(6): 2014-2044. |
| [22] | Jez JM. Structural biology of plant sulfur metabolism: from sulfate to glutathione [J]. J Exp Bot, 2019, 70(16): 4089-4103. |
| [23] | Tang WL, Zhao YJ, Zeng JJ, et al. Integration of small RNA and transcriptome sequencing reveal the roles of miR395 and ATP sulfurylase in developing seeds of Chinese kale [J]. Front Plant Sci, 2022, 12: 778848. |
| [24] | Xu XP, Yang CL, Zheng YP, et al. Transcriptome-wide identification of miRNAs and their targets during riboflavin-promoted dormancy release in Lilium 'Siberia' [J]. Horticulturae, 2025, 11(1): 17. |
| [25] | Xu SJ, Chen RZ, Zhang XQ, et al. The evolutionary tale of lilies: giant genomes derived from transposon insertions and polyploidization [J]. Innov, 2024, 5(6): 100726. |
| [26] | 孙红梅, 李天来, 李云飞. 低温解除休眠过程中兰州百合鳞茎酚类物质含量及相关酶活性变化 [J]. 中国农业科学, 2004, 37(11): 1777-1782. |
| Sun HM, Li TL, Li YF. Changes of phenols content and activity of enzymes related to phenols in lily bulbs stored at different cold temperatures for breaking dormancy [J]. Sci Agric Sin, 2004, 37(11): 1777-1782. | |
| [27] | 梁云, 袁素霞, 冯慧颖, 等. 百合肌动蛋白基因lilyActin的克隆与表达分析 [J]. 园艺学报, 2013, 40(7): 1318-1326. |
| Liang Y, Yuan SX, Feng HY, et al. Cloning and expression analysis of actin gene (lilyActin) from lily [J]. Acta Hortic Sin, 2013, 40(7): 1318-1326. | |
| [28] | Hesse H, Nikiforova V, Gakière B, et al. Molecular analysis and control of cysteine biosynthesis: integration of nitrogen and sulphur metabolism [J]. J Exp Bot, 2004, 55(401): 1283-1292. |
| [29] | Zhang HM, Lang ZB, Zhu JK. Dynamics and function of DNA methylation in plants [J]. Nat Rev Mol Cell Biol, 2018, 19(8): 489-506. |
| [30] | Lu L, Chen XS, Sanders D, et al. High-resolution mapping of H4K16 and H3K23 acetylation reveals conserved and unique distribution patterns in Arabidopsis and rice [J]. Epigenetics, 2015, 10(11): 1044-1053. |
| [31] | Luo CY, Sidote DJ, Zhang Y, et al. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production [J]. Plant J, 2013, 73(1): 77-90. |
| [32] | Yang ZY, Hui SG, Lv Y, et al. miR395-regulated sulfate metabolism exploits pathogen sensitivity to sulfate to boost immunity in rice [J]. Mol Plant, 2022, 15(4): 671-688. |
| [33] | Rajjou L, Duval M, Gallardo K, et al. Seed germination and vigor [J]. Annu Rev Plant Biol, 2012, 63: 507-533. |
| [34] | Akbudak MA, Filiz E. Genome-wide analyses of ATP sulfurylase (ATPS) genes in higher plants and expression profiles in sorghum (Sorghum bicolor) under cadmium and salinity stresses [J]. Genomics, 2019, 111(4): 579-589. |
| [35] | Kim WS, Sun-Hyung J, Oehrle NW, et al. Overexpression of ATP sulfurylase improves the sulfur amino acid content, enhances the accumulation of Bowman-Birk protease inhibitor and suppresses the accumulation of the β-subunit of β-conglycinin in soybean seeds [J]. Sci Rep, 2020, 10(1): 14989. |
| [36] | Yatusevich R, Mugford SG, Matthewman C, et al. Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana [J]. Plant J, 2010, 62(1): 1-11. |
| [37] | Matthewman C. Arabidopsis ATP sulfurylase: roles and regulation of individual isoforms [D]. Norwich: University of East Anglia, 2010. |
| [1] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [2] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [3] | ZHANG Yong-yan, GUO Si-jian, LI Jing, HAO Si-yi, LI Rui-de, LIU Jia-peng, CHENG Chun-zhen. Gene Cloning and Functional Analysis of the Anthocyanin-related VcGSTF19 Gene in Blueberry (Vaccinium corymbosum L.) [J]. Biotechnology Bulletin, 2025, 41(9): 139-146. |
| [4] | LA Gui-xiao, ZHAO Yu-long, DAI Dan-dan, YU Yong-liang, GUO Hong-xia, SHI Gui-xia, JIA Hui, YANG Tie-gang. Identification of Plasma Membrane H+-ATPase Gene Family in Safflower and Expression Analysis in Response to Low Nitrogen and Low Phosphorus Stress [J]. Biotechnology Bulletin, 2025, 41(8): 220-233. |
| [5] | LI Kai-jie, WU Yao, LI Dan-dan. Cloning of Gene CtbHLH128 in Safflower and Response Function Regulating Drought Stress [J]. Biotechnology Bulletin, 2025, 41(8): 234-241. |
| [6] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [7] | KANG Qin, WANG Xia, SHEN Ming-yang, XU Jing-tian, CHEN Shi-lan, LIAO Ping-yang, XU Wen-zhi, WU Wei, XU Dong-bei. Cloning and Expression Analysis of the UV-B Receptor Gene McUVR8 in Mentha canadensis L. [J]. Biotechnology Bulletin, 2025, 41(8): 255-266. |
| [8] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [9] | LI Xin-ni, LI Jun-yi, MA Xue-hua, HE Wei, LI Jia-li, YU Jia, CAO Xiao-ning, QIAO Zhi-jun, LIU Si-chen. Identification of the PMEI Gene Family of Pectin Methylesterase Inhibitor in Foxtail Millet and Analysis of Its Response to Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(7): 150-163. |
| [10] | ZHANG Yong, SONG Sheng-long, LI Yong-tai, ZHANG Xin-yu, LI Yan-jun. Cloning of GhSWEET9 in Upland Cotton and Functional Analysis of Resistance to Verticillium Wilt [J]. Biotechnology Bulletin, 2025, 41(6): 144-154. |
| [11] | PEI Jing-qi, ZHAO Meng-ran, HUANG Chen-yang, WU Xiang-li. Discovery and Verification of a Functional Gene Influencing the Growth and Development of Pleurotus ostreatus [J]. Biotechnology Bulletin, 2025, 41(6): 327-334. |
| [12] | XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1 [J]. Biotechnology Bulletin, 2025, 41(6): 119-129. |
| [13] | CHENG Shan, WANG Hui-wei, CHEN Chen, ZHU Ya-jing, LI Chun-xin, BIE Hai, WANG Shu-feng, CHEN Xian-gong, ZHANG Xiang-ge. Cloning of MYB Transcription Factor Gene CeMYB154 and Analysis of Salt Tolerance Function in Cyperus esculentus [J]. Biotechnology Bulletin, 2025, 41(6): 218-228. |
| [14] | LIU Xin, WANG Jia-wen, LI Jin-wei, MOU Ce, YANG Pan-pan, MING Jun, XU Lei-feng. Cloning and Expression Analysis of Three LdBBXs in Lilium davidii var. willmottiae [J]. Biotechnology Bulletin, 2025, 41(5): 186-196. |
| [15] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||