








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (5): 186-196.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1174
LIU Xin1,2( ), WANG Jia-wen2, LI Jin-wei2,3, MOU Ce1,2, YANG Pan-pan2, MING Jun2, XU Lei-feng2(
), WANG Jia-wen2, LI Jin-wei2,3, MOU Ce1,2, YANG Pan-pan2, MING Jun2, XU Lei-feng2( )
)
Received:2024-12-05
Online:2025-05-26
Published:2025-06-05
Contact:
XU Lei-feng
E-mail:14763795646@163.com;xuleifeng@caas.cn
LIU Xin, WANG Jia-wen, LI Jin-wei, MOU Ce, YANG Pan-pan, MING Jun, XU Lei-feng. Cloning and Expression Analysis of Three LdBBXs in Lilium davidii var. willmottiae[J]. Biotechnology Bulletin, 2025, 41(5): 186-196.
| 引物 Primer | 序列 Sequence (5′‒3′) | 退火温度 Annealing temperature (℃) |
|---|---|---|
| LdBBX21-F | GGAGCCAGATCGGAGAG | 55.5 |
| LdBBX21-R | GCTTGAGGAAACAGAAAGAGG | |
| LdBBX22-F | GAGAGAGAGGAGAGAGAGA | 51.5 |
| LdBBX22-R | TCGACCCATAATAACACATC | |
| LdBBX24-F | GGGGATTGTCCAACTATATCCC | 54.8 |
| LdBBX24-R | TTCTCCGTTCATGTTTTGTGC | |
| LdBBX21-qRT-F | CTCCATCCACAGGGCCAAC | 59.0 |
| LdBBX21-qRT-R | CAAACCGTCGAGGATGAGACG | |
| LdBBX22-qRT-F | GGAGGCGACGGTGATGTG | 60.0 |
| LdBBX22-qRT-R | CGGAAGCGTTGCCAGAGATT | |
| LdBBX24-qRT-F | CAACACAGCAGTCGCCTTC | 57.5 |
| LdBBX24-qRT-R | CCACCAGAAAACTCCTTCTGC | |
| LilyActin-F | GCACCTGAAGAGCACCCT | 60.0 |
| LilyActin-R | TGGCGTAAAGCGACAAAA | |
| LdBBX21-2300-F | ATTTGGAGAGGACAGGGTACGGAGCCAGATCGGAGAG | 55.5 |
| LdBBX21-2300-R | CACCATGGTACTAGTGTCGAGCTTGAGGAAACAGAAAGAGG | |
| LdBBX22-2300-F | ATTTGGAGAGGACAGGGTAC AGAGAGAGATCTTGTCGGC | 55.0 |
| LdBBX22-2300-R | CACCATGGTACTAGTGTCGACATACTGATAGACATGTACAGCAG | |
| LdBBX24-2300-F | ATTTGGAGAGGACAGGGTACGGGGATTGTCCAACTATATCCC | 54.8 |
| LdBBX24-2300-R | CACCATGGTACTAGTGTCGATTCTCCGTTCATGTTTTGTGC | |
| Detect-2300-F | ATTTGGAGAGGACAGGGTAC | 55.0 |
| Detect-2300-R | CACCATGGTACTAGTGTCGA |
Table 1 Information of the primers used in this study
| 引物 Primer | 序列 Sequence (5′‒3′) | 退火温度 Annealing temperature (℃) |
|---|---|---|
| LdBBX21-F | GGAGCCAGATCGGAGAG | 55.5 |
| LdBBX21-R | GCTTGAGGAAACAGAAAGAGG | |
| LdBBX22-F | GAGAGAGAGGAGAGAGAGA | 51.5 |
| LdBBX22-R | TCGACCCATAATAACACATC | |
| LdBBX24-F | GGGGATTGTCCAACTATATCCC | 54.8 |
| LdBBX24-R | TTCTCCGTTCATGTTTTGTGC | |
| LdBBX21-qRT-F | CTCCATCCACAGGGCCAAC | 59.0 |
| LdBBX21-qRT-R | CAAACCGTCGAGGATGAGACG | |
| LdBBX22-qRT-F | GGAGGCGACGGTGATGTG | 60.0 |
| LdBBX22-qRT-R | CGGAAGCGTTGCCAGAGATT | |
| LdBBX24-qRT-F | CAACACAGCAGTCGCCTTC | 57.5 |
| LdBBX24-qRT-R | CCACCAGAAAACTCCTTCTGC | |
| LilyActin-F | GCACCTGAAGAGCACCCT | 60.0 |
| LilyActin-R | TGGCGTAAAGCGACAAAA | |
| LdBBX21-2300-F | ATTTGGAGAGGACAGGGTACGGAGCCAGATCGGAGAG | 55.5 |
| LdBBX21-2300-R | CACCATGGTACTAGTGTCGAGCTTGAGGAAACAGAAAGAGG | |
| LdBBX22-2300-F | ATTTGGAGAGGACAGGGTAC AGAGAGAGATCTTGTCGGC | 55.0 |
| LdBBX22-2300-R | CACCATGGTACTAGTGTCGACATACTGATAGACATGTACAGCAG | |
| LdBBX24-2300-F | ATTTGGAGAGGACAGGGTACGGGGATTGTCCAACTATATCCC | 54.8 |
| LdBBX24-2300-R | CACCATGGTACTAGTGTCGATTCTCCGTTCATGTTTTGTGC | |
| Detect-2300-F | ATTTGGAGAGGACAGGGTAC | 55.0 |
| Detect-2300-R | CACCATGGTACTAGTGTCGA |

Fig. 2 LdBBX21, LdBBX22, and LdBBX24 amplification electropherogram and their translated amino acid sequencesA, C, E: Amplification electropherogram of LdBBX21, LdBBX22, LdBBX24; B, D, F: CDS sequence of LdBBX21, LdBBX22, LdBBX24 and its translated amino acid sequence
| 蛋白 Protein | 分子式 Molecular formula | 分子量 Molecular weight (kD) | 等电点 pI | 不稳定系数 Instability index |
|---|---|---|---|---|
| LdBBX21 | C948H1477N273O303S14 | 22.00 | 5.87 | 60.39 |
| LdBBX22 | C1425H2185N393O456S18 | 32.70 | 5.28 | 61.55 |
| LdBBX24 | C1151H1792N312O357S19 | 26.32 | 4.91 | 60.48 |
Table 2 Basic physicochemical properties of LdBBX21, LdBBX22, and LdBBX24 proteins
| 蛋白 Protein | 分子式 Molecular formula | 分子量 Molecular weight (kD) | 等电点 pI | 不稳定系数 Instability index |
|---|---|---|---|---|
| LdBBX21 | C948H1477N273O303S14 | 22.00 | 5.87 | 60.39 |
| LdBBX22 | C1425H2185N393O456S18 | 32.70 | 5.28 | 61.55 |
| LdBBX24 | C1151H1792N312O357S19 | 26.32 | 4.91 | 60.48 |
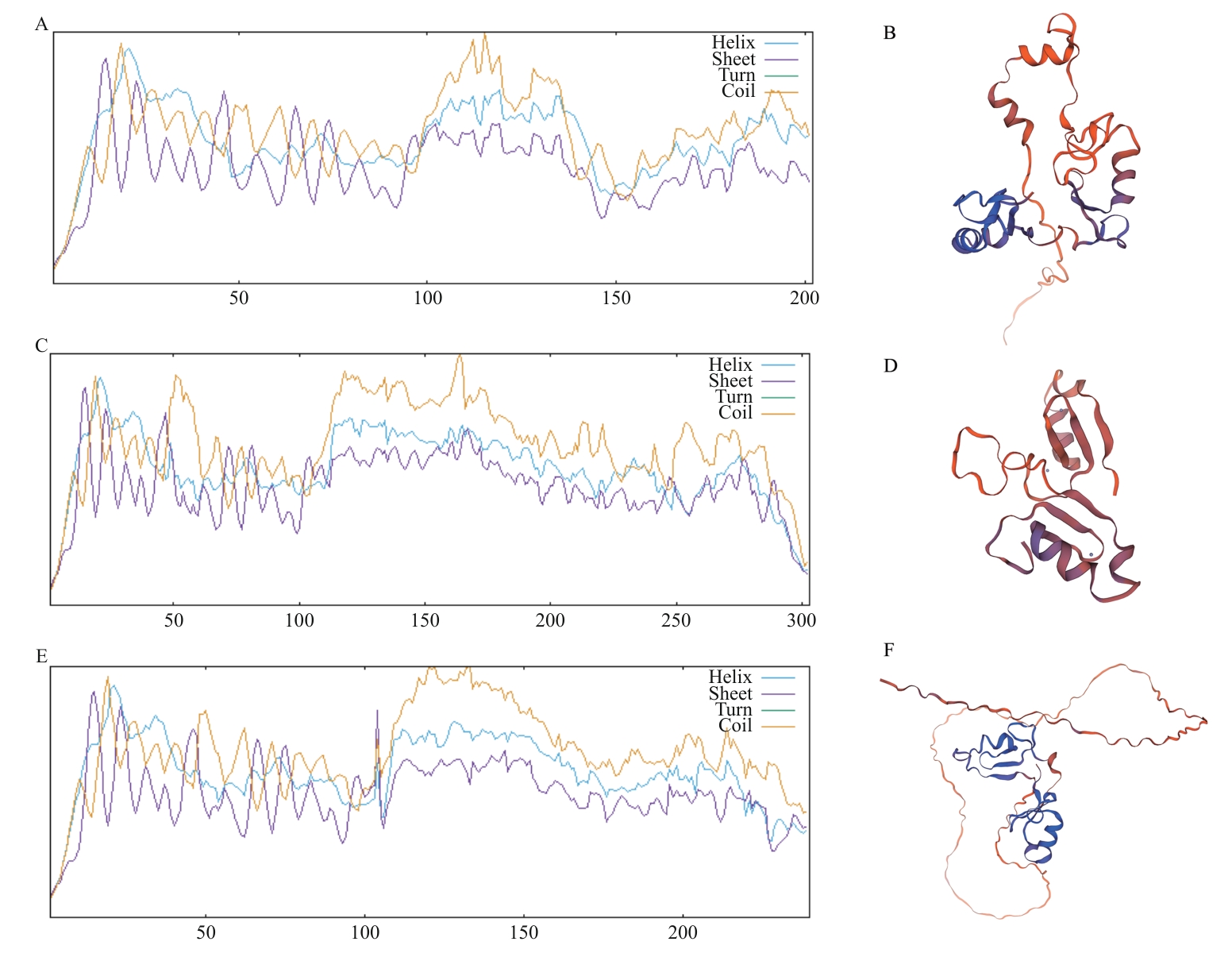
Fig. 4 Prediction of secondary and tertiary structures of LdBBX21, LdBBX22, and LdBBX24 proteinsA, C, E: Secondary structure prediction of LdBBX21, LdBBX22, and LdBBX24 protein. B, D, F: Tertiary structure prediction of LdBBX21, LdBBX22, and LdBBX24 protein
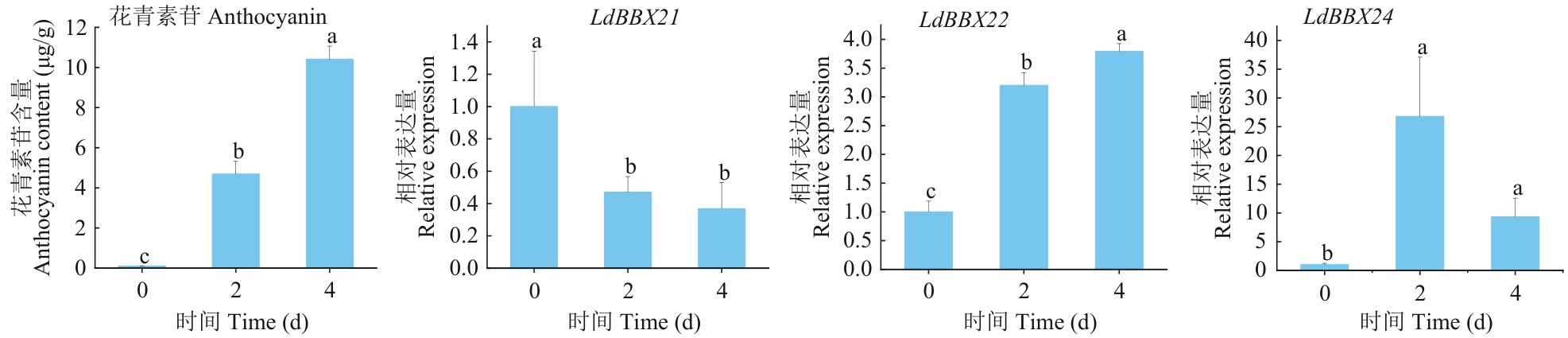
Fig. 9 Relative expressions and anthocyanin contents of LdBBX21, LdBBX22, and LdBBX24 genes under light treatment for 0, 2, and 4 dDifferent letters above columns indicate significant differences at P<0.05 level. The same below
| 1 | 徐丽, 朱田田, 晋玲, 等. 基于感官评价和化学计量学的兰州百合品质分析 [J]. 食品工业科技, 2024, 45(18): 219-227. |
| Xu L, Zhu TT, Jin L, et al. Comprehensive evaluation of Lilium davidii var. willmottiae (E. H. Wilson) Raffill quality based on sensory evaluation and chemometrics [J]. Sci Technol Food Ind, 2024, 45(18): 219-227. | |
| 2 | 杨慧勤, 王佳丽, 李思蕤, 等. 茄科蔬菜花青素苷分子调控研究进展 [J]. 生物工程学报, 2022, 38(5): 1738-1752. |
| Yang HQ, Wang JL, Li SR, et al. Advances in the molecular regulation of anthocyanins in solanaceous vegetables [J]. Chin J Biotechnol, 2022, 38(5): 1738-1752. | |
| 3 | 范文广, 柴佳靖, 李保豫, 等. 百合花青苷分子调控研究进展 [J]. 植物遗传资源学报, 2023, 24(5): 1236-1247. |
| Fan WG, Chai JJ, Li BY, et al. Advances in molecular regulation of anthocyanin biosynthesis in Lilium [J]. J Plant Genet Resour, 2023, 24(5): 1236-1247. | |
| 4 | Fan WG, Bai P, Chen RR, et al. Postharvest light irradiation induces anthocyanin accumulation in fresh-cut lily Bulb (Lilium davidii var. unicolor) scales [J]. J Food Process Preserv, 2024, 2024(1): 7984106. |
| 5 | 毕蒙蒙, 曹雨薇, 宋蒙, 等. 百合花色研究进展 [J]. 园艺学报, 2021, 48(10): 2073-2086. |
| Bi MM, Cao YW, Song M, et al. Advances in flower color research of Lilium [J]. Acta Hortic Sin, 2021, 48(10): 2073-2086. | |
| 6 | 路喻丹, 刘晓驰, 冯新, 等. 猕猴桃BBX基因家族成员鉴定与转录特征分析 [J]. 生物技术通报, 2024, 40(2): 172-182. |
| Lu YD, Liu XC, Feng X, et al. Identification of the kiwifruit BBX gene family and analysis of their transcriptional characteristics [J]. Biotechnol Bull, 2024, 40(2): 172-182. | |
| 7 | Gangappa SN, Botto JF. The BBX family of plant transcription factors [J]. Trends Plant Sci, 2014, 19(7): 460-470. |
| 8 | Zou ZY, Wang RH, Wang R, et al. Genome-wide identification, phylogenetic analysis, and expression profiling of the BBX family genes in pear [J]. J Hortic Sci Biotechnol, 2018, 93(1): 37-50. |
| 9 | Yadav A, Ravindran N, Singh D, et al. Role of Arabidopsis BBX proteins in light signaling [J]. J Plant Biochem Biotechnol, 2020, 29(4): 623-635. |
| 10 | Bursch K, Niemann ET, Nelson DC, et al. Karrikins control seedling photomorphogenesis and anthocyanin biosynthesis through a HY5-BBX transcriptional module [J]. Plant J, 2021, 107(5): 1346-1362. |
| 11 | Yadukrishnan P, Job N, Johansson H, et al. Opposite roles of group IV BBX proteins: exploring missing links between structural and functional diversity [J]. Plant Signal Behav, 2018: e1462641. |
| 12 | Sarmiento F. The BBX subfamily IV: additional cogs and sprockets to fine-tune light-dependent development [J]. Plant Signal Behav, 2013, 8(4): e23831. |
| 13 | Cao J, Yuan JL, Zhang YL, et al. Multi-layered roles of BBX proteins in plant growth and development [J]. Stress Biol, 2023, 3(1): 1. |
| 14 | Bursch K, Toledo-Ortiz G, Pireyre M, et al. Identification of BBX proteins as rate-limiting cofactors of HY5 [J]. Nat Plants, 2020, 6(8): 921-928. |
| 15 | Gangappa SN, Crocco CD, Johansson H, et al. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis [J]. Plant Cell, 2013, 25(4): 1243-1257. |
| 16 | Xu DQ, Jiang Y, Li J, et al. The B-box domain protein BBX21 promotes photomorphogenesis [J]. Plant Physiol, 2018, 176(3): 2365-2375. |
| 17 | Crocco CD, Holm M, Yanovsky MJ, et al. AtBBX21 and COP1 genetically interact in the regulation of shade avoidance [J]. Plant J, 2010, 64(4): 551-562. |
| 18 | Podolec R, Wagnon TB, Leonardelli M, et al. Arabidopsis B-box transcription factors BBX20-22 promote UVR8 photoreceptor-mediated UV-B responses [J]. Plant J, 2022, 111(2): 422-439. |
| 19 | Zhang B, Zhu ZZ, Qu D, et al. MdBBX21, a B-box protein, positively regulates light-induced anthocyanin accumulation in apple peel [J]. Front Plant Sci, 2021, 12: 774446. |
| 20 | Xie YX, Miao TT, Lyu SH, et al. Arabidopsis ERD15 regulated by BBX24 plays a positive role in UV-B signaling [J]. Plant Sci, 2024, 343: 112077. |
| 21 | Singh A, Ram H, Abbas N, et al. Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana [J]. J Biol Chem, 2012, 287(31): 25995-26009. |
| 22 | Job N, Yadukrishnan P, Bursch K, et al. Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains [J]. Plant Physiol, 2018, 176(4): 2963-2976. |
| 23 | Jakoby M, Weisshaar B, Dröge-Laser W, et al. bZIP transcription factors in Arabidopsis [J]. Trends Plant Sci, 2002, 7(3): 106-111. |
| 24 | Bai SL, Saito T, Honda C, et al. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis [J]. Planta, 2014, 240(5): 1051-1062. |
| 25 | Fang HC, Dong YH, Yue XX, et al. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature [J]. Plant Cell Environ, 2019, 42(7): 2090-2104. |
| 26 | An JP, Wang XF, Zhang XW, et al. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation [J]. Plant Biotechnol J, 2019, 17(12): 2231-2233. |
| 27 | Zhang B, Yang HJ, Qu D, et al. The MdBBX22-miR858-MdMYB9/11/12 module regulates proanthocyanidin biosynthesis in apple peel [J]. Plant Biotechnol J, 2022, 20(9): 1683-1700. |
| 28 | Bai SL, Tao RY, Yin L, et al. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit [J]. Plant J, 2019, 100(6): 1208-1223. |
| 29 | 曹雨薇, 徐雷锋, 杨盼盼, 等. 百合花青素苷呈色类型中3种R2R3-MYBs基因的差异表达 [J]. 园艺学报, 2019, 46(5): 955-963. |
| Cao YW, Xu LF, Yang PP, et al. Differential expression of three R2R3-MYBs genes regulating anthocyanin pigmentation patterns in Lilium spp [J]. Acta Hortic Sin, 2019, 46(5): 955-963. | |
| 30 | Chang CJ, Maloof JN, Wu SH. COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis [J]. Plant Physiol, 2011, 156(1): 228-239. |
| 31 | Xu DQ, Jiang Y, Li JG, et al. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation [J]. Proc Natl Acad Sci U S A, 2016, 113(27): 7655-7660. |
| 32 | Lyu GZ, Li DB, Li SS. Bioinformatics analysis of BBX family genes and its response to UV-B in Arabidopsis thaliana [J]. Plant Signal Behav, 2020, 15(9): 1782647. |
| 33 | Huang YW, Xiong H, Xie YX, et al. BBX24 interacts with DELLA to regulate UV-B-induced photomorphogenesis in Arabidopsis thaliana [J]. Int J Mol Sci, 2022, 23(13): 7386. |
| 34 | Gangappa SN, Holm M, Botto JF. Molecular interactions of BBX24 and BBX25 with HYH, HY5 HOMOLOG, to modulate Arabidopsis seedling development [J]. Plant Signal Behav, 2013, 8(8): e25208. |
| 35 | Hao XL, Zhong YJ, Tzmann HN, et al. Light-induced artemisinin biosynthesis is regulated by the bZIP transcription factor AaHY5 in Artemisia annua [J]. Plant Cell Physiol, 2019, 60(8): 1747-1760. |
| 36 | He WZ, Liu H, Wu Z, et al. The AaBBX21-AaHY5 module mediates light-regulated artemisinin biosynthesis in Artemisia annua L [J]. J Integr Plant Biol, 2024, 66(8): 1735-1751. |
| 37 | Yang GQ, Zhang CL, Dong HX, et al. Activation and negative feedback regulation of SlHY5 transcription by the SlBBX20/21-SlHY5 transcription factor module in UV-B signaling [J]. Plant Cell, 2022, 34(5): 2038-2055. |
| 38 | Li SR, Ou CQ, Wang F, et al. Ppbbx24-del mutant positively regulates light-induced anthocyanin accumulation in the ‘Red Zaosu’ pear (Pyrus pyrifolia White Pear Group) [J]. J Integr Agric, 2024. . |
| [1] | LI Zhi-qiang, LUO Zheng-qian, XU Lin-li, ZHOU Guo-hui, QU Si-yu, LIU En-liang, XU Dong-ting. Identification of the Soybean R2R3-MYB Gene Family Based on the T2T Genome and Their Expression Analysis under Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 141-152. |
| [2] | ZHAO Jing, GUO Qian, LI Rui-qi, LEI Ying-yang, YUE Ai-qin, ZHAO Jin-zhong, YIN Cong-cong, DU Wei-jun, NIU Jing-ping. Sequence Analysis and Induced Expression Analysis of GmGST Gene Cluster Genes in Soybean [J]. Biotechnology Bulletin, 2025, 41(5): 129-140. |
| [3] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [4] | SUN Tian-guo, YI Lan, QIN Xu-yang, QIAO Meng-xue, GU Xin-ying, HAN Yi, SHA Wei, ZHANG Mei-juan, MA Tian-yi. Genome-wide Identification of the DABB Gene Family in Brassica rapa ssp. pekinensis and Expression Analysis under Saline and Alkali Stress [J]. Biotechnology Bulletin, 2025, 41(4): 156-165. |
| [5] | WANG Tian-tian, CHANG Xue-rui, HUANG Wan-yang, HUANG Jia-xin, MIAO Ru-yi, LIANG Yan-ping, WANG Jing. Identification and Analysis of GASA Gene Family in Pepper (Capsicum annuum L.) [J]. Biotechnology Bulletin, 2025, 41(4): 166-175. |
| [6] | HUANG Jin-heng, HUANG Xi, ZHANG Jia-yan, ZHOU Xin-yu, LIAO Pei-ran, YANG Quan. Identification and Expression Analysis of the C3H Gene Family in Grona styracifolia across Different Varieties [J]. Biotechnology Bulletin, 2025, 41(4): 243-255. |
| [7] | BAN Qiu-yan, ZHAO Xin-yue, CHI Wen-jing, LI Jun-sheng, WANG Qiong, XIA Yao, LIANG Li-yun, HE Wei, LI Ye-yun, ZHAO Guang-shan. Cloning of Phytochrome Interaction Factor CsPIF3a and Its Response to Light and Temperature Stress in Camellia sinensis [J]. Biotechnology Bulletin, 2025, 41(4): 256-265. |
| [8] | WANG Chen, LIU Guo-mei, CHEN Chang, ZHANG Jin-long, YAO Lin, SUN Xuan, DU Chun-fang. Genome-wide Identification and Expression Analysis of CCDs Family in Brassia rapa L. [J]. Biotechnology Bulletin, 2025, 41(3): 161-170. |
| [9] | PENG Ting, LIN Ying, TAN Yuan-yuan, RAO Ying, HUANG Qin, ZHANG Wen-e, WANG Bo, TIAN Rui-feng, LIU Guo-feng. Cloning and Expression Analysis of AwANS Genes in Allium wallichii [J]. Biotechnology Bulletin, 2025, 41(3): 230-239. |
| [10] | MA Tian-yi, XU Jia-jia, LU Wen-jing, WU Yan, SHA Wei, ZHANG Mei-juan, PENG Yi-fang. Expression Analysis and Resistance Identification of BrcGASA3 in Chinese Cabbage ‘Jinxiaotong’ Cultivar under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 127-138. |
| [11] | XU Yuan-meng, MAO Jiao, WANG Meng-yao, WANG Shu, REN Jiang-ling, LIU Yu-han, LIU Si-chen, QIAO Zhi-jun, WANG Rui-yun, CAO Xiao-ning. Cloning and Expression Characteristics Analysis of Millet Genes PmDEP1 and PmEP3 [J]. Biotechnology Bulletin, 2025, 41(2): 150-162. |
| [12] | JIA Zi-jian, WANG Bao-qiang, CHEN Li-fei, WANG Yi-zhen, WEI Xiao-hong, ZHAO Ying. Expression Patterns of CHX Gene Family in Quinoa in Response to NO under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 163-174. |
| [13] | JIAO Xiao-yu, WU Qiong, LIU Dan-dan, SUN Ming-hui, RUAN Xu, WANG Lei-gang, WANG Wen-jie. Cloning and Functional Analysis of CsWAK8 Gene from Camellia sinensis during Cold Stress [J]. Biotechnology Bulletin, 2025, 41(2): 210-220. |
| [14] | QIAN Zheng-yi, WU Shao-fang, CAO Shu-yi, SONG Ya-xin, PAN Xin-feng, LI Zhao-wei, FAN Kai. Identification of the NAC Transcription Factors in Nymphaea colorata and Their Expression Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 234-247. |
| [15] | XIANG Chun-fan, LI Le-song, WANG Juan, LIANG Yan-li, YANG Sheng-chao, LI Meng-fei, ZHAO Yan. Functional Identification and Expression Analysis of Cinnamonyl Alcohol Dehydrogenase AsCAD in Angelica sinensis [J]. Biotechnology Bulletin, 2025, 41(2): 295-308. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||