








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (10): 186-195.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0421
Previous Articles Next Articles
WENG Hui-ting1,2( ), GUO Hui-ming3, CHENG Hong-mei3, LI Jun2, ZHANG Chao2, LIU Hai-yang1(
), GUO Hui-ming3, CHENG Hong-mei3, LI Jun2, ZHANG Chao2, LIU Hai-yang1( ), SU Xiao-feng3(
), SU Xiao-feng3( )
)
Received:2025-04-22
Online:2025-10-26
Published:2025-10-28
Contact:
LIU Hai-yang, SU Xiao-feng
E-mail:wht_bio@163.com;liuhaiyang001@163.com;suxiaofeng@caas.cn
WENG Hui-ting, GUO Hui-ming, CHENG Hong-mei, LI Jun, ZHANG Chao, LIU Hai-yang, SU Xiao-feng. Establishment and Application of Droplet Digital PCR Detection Methods for Four Major Verticillium Wilt Pathogens[J]. Biotechnology Bulletin, 2025, 41(10): 186-195.
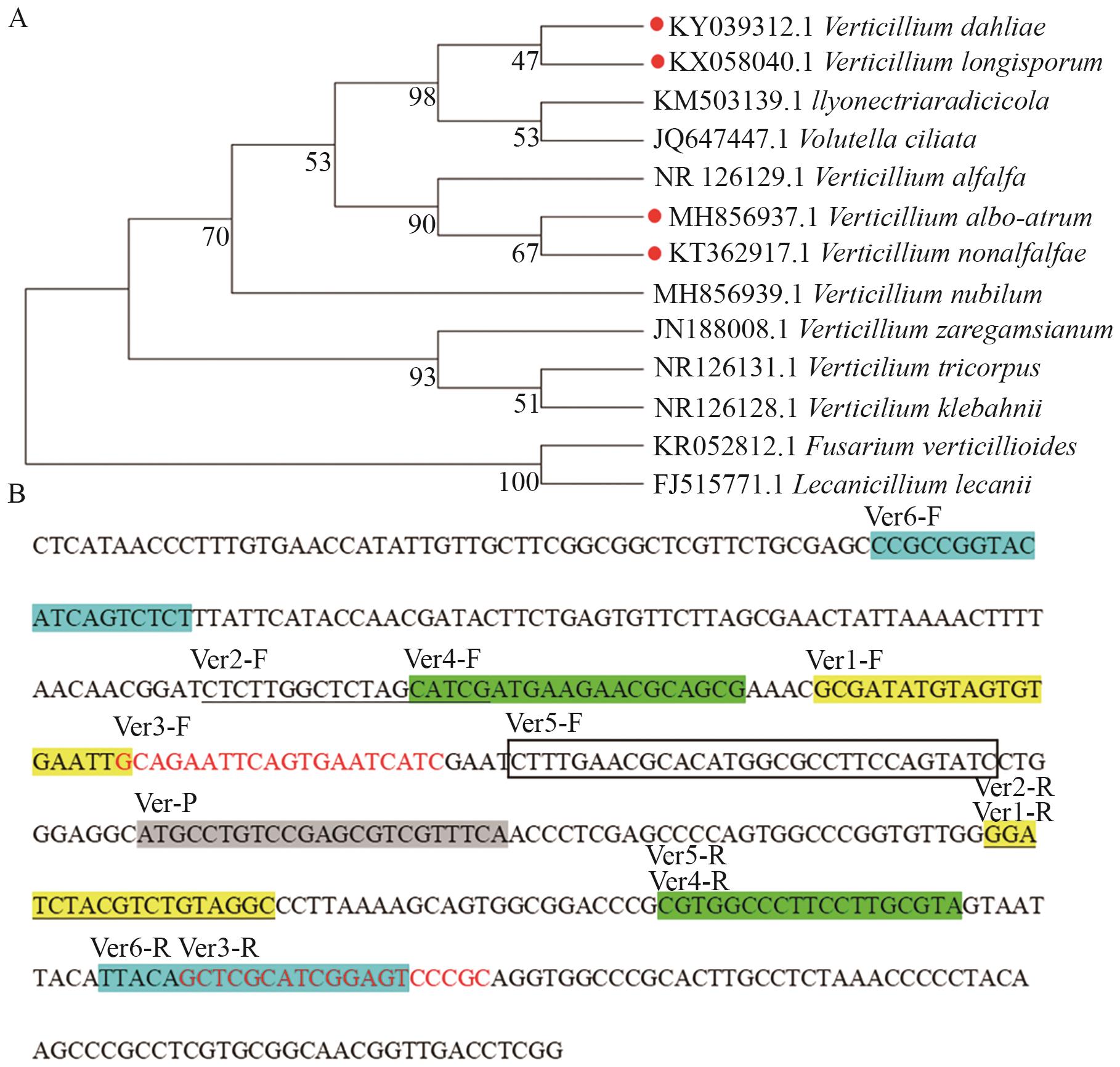
Fig. 1 Phylogenetic analysis and the position of primers and probes in ITS sequencesA: Construction of the phylogenetic tree for Verticillium and other fungi; B: position of primers and probes in ITS sequence
引物名称 Primer name | 序列 Sequence (5′-3′) | 长度 Length (bp) |
|---|---|---|
| Ver1 | F:GCGATATGTAGTGTGAATTG | 152 |
| R:GCCTACAGACGTAGATCC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver2 | F:CTCTTGGCTCTAGCATCG | 189 |
| R:GCCTACAGACGTAGATCC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver3 | F:GCAGAATTCAGTGAATCATC | 203 |
| R:GCGGGACTCCGATGCGAGC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver4 | F:CATCGATGAAGAACGCAGCG | 97 |
| R:TACGCAAGGAAGGGCCACG | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver5 | F:CATCGATGAAGAACGCAGCG | 218 |
| R:TACGCAAGGAAGGGCCACG | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver6 | F:CCGCCGGTACATCAGTCTCT | 337 |
| R:ACTCCGATGCGAGCTGTAA | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA |
Table 1 Primers and probes used in this study
引物名称 Primer name | 序列 Sequence (5′-3′) | 长度 Length (bp) |
|---|---|---|
| Ver1 | F:GCGATATGTAGTGTGAATTG | 152 |
| R:GCCTACAGACGTAGATCC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver2 | F:CTCTTGGCTCTAGCATCG | 189 |
| R:GCCTACAGACGTAGATCC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver3 | F:GCAGAATTCAGTGAATCATC | 203 |
| R:GCGGGACTCCGATGCGAGC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver4 | F:CATCGATGAAGAACGCAGCG | 97 |
| R:TACGCAAGGAAGGGCCACG | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver5 | F:CATCGATGAAGAACGCAGCG | 218 |
| R:TACGCAAGGAAGGGCCACG | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver6 | F:CCGCCGGTACATCAGTCTCT | 337 |
| R:ACTCCGATGCGAGCTGTAA | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA |
菌株DNA Strain DNA | DNA浓度 DNA concentration (ng/μL) | Ver1 | Ver2 | Ver3 | Ver4 | Ver5 | Ver6 |
|---|---|---|---|---|---|---|---|
| Vd | 18.2 | 19.67 | 17.70 | 18.28 | 18.63 | 15.02 | 16.24 |
| Vl | 19 | 15.65 | 13.53 | 14.42 | 14.53 | 11.19 | 11.93 |
| Vna | 22.2 | 17.96 | 16.04 | 16.70 | 16.99 | 13.48 | 14.25 |
| Vaa | 23.8 | 17.19 | 15.14 | 15.85 | 16.13 | 12.49 | 14.07 |
阴性对照 Negative control | 0 | Un. | 37 | 37 | Un. | 37 | 37 |
Table 2 Ct values in different sets of primers and probes
菌株DNA Strain DNA | DNA浓度 DNA concentration (ng/μL) | Ver1 | Ver2 | Ver3 | Ver4 | Ver5 | Ver6 |
|---|---|---|---|---|---|---|---|
| Vd | 18.2 | 19.67 | 17.70 | 18.28 | 18.63 | 15.02 | 16.24 |
| Vl | 19 | 15.65 | 13.53 | 14.42 | 14.53 | 11.19 | 11.93 |
| Vna | 22.2 | 17.96 | 16.04 | 16.70 | 16.99 | 13.48 | 14.25 |
| Vaa | 23.8 | 17.19 | 15.14 | 15.85 | 16.13 | 12.49 | 14.07 |
阴性对照 Negative control | 0 | Un. | 37 | 37 | Un. | 37 | 37 |
植物病原体 Plant pathogen | Ct值 Ct value | ||||
|---|---|---|---|---|---|
| Ver2 | Ver3 | Ver5 | Ver6 | ||
稻瘟病菌 Magnaporthe oryzae | Un. | Un. | Un. | Un. | |
玉米小斑病菌 Bipolaris maydis | Un. | Un. | Un. | Un. | |
玉米大斑病菌 Exserohilum turcicum | Un. | Un. | Un. | Un. | |
尖孢镰刀菌粘团专化型 Fusarium fujikuroi f. sp. nirenbergiae | Un. | Un. | Un. | Un. | |
禾谷丝核菌 Rhizoctonia solani | Un. | Un. | Un. | Un. | |
南方根结线虫 Meloidogyne incognita | Un. | Un. | Un. | Un. | |
假禾谷镰孢菌 Fusarium pseudograminearum | Un. | Un. | Un. | Un. | |
水稻稻曲病菌 Ustilaginoidea virens | Un. | Un. | Un. | Un. | |
西瓜嗜酸菌 Acidovorax citrulli | Un. | Un. | Un. | Un. | |
水稻白叶枯病菌 Xanthomonas oryzae pv. oryzae | Un. | Un. | Un. | Un. | |
丁香假单胞菌 Pseudomonas syringae | Un. | Un. | Un. | Un. | |
青枯菌 Ralstonia solanacearum | Un. | Un. | Un. | Un. | |
野油菜黄单胞菌 Xanthomonas campestris pv. campestris | Un. | Un. | Un. | Un. | |
Table 3 Specific analysis of primers/probe
植物病原体 Plant pathogen | Ct值 Ct value | ||||
|---|---|---|---|---|---|
| Ver2 | Ver3 | Ver5 | Ver6 | ||
稻瘟病菌 Magnaporthe oryzae | Un. | Un. | Un. | Un. | |
玉米小斑病菌 Bipolaris maydis | Un. | Un. | Un. | Un. | |
玉米大斑病菌 Exserohilum turcicum | Un. | Un. | Un. | Un. | |
尖孢镰刀菌粘团专化型 Fusarium fujikuroi f. sp. nirenbergiae | Un. | Un. | Un. | Un. | |
禾谷丝核菌 Rhizoctonia solani | Un. | Un. | Un. | Un. | |
南方根结线虫 Meloidogyne incognita | Un. | Un. | Un. | Un. | |
假禾谷镰孢菌 Fusarium pseudograminearum | Un. | Un. | Un. | Un. | |
水稻稻曲病菌 Ustilaginoidea virens | Un. | Un. | Un. | Un. | |
西瓜嗜酸菌 Acidovorax citrulli | Un. | Un. | Un. | Un. | |
水稻白叶枯病菌 Xanthomonas oryzae pv. oryzae | Un. | Un. | Un. | Un. | |
丁香假单胞菌 Pseudomonas syringae | Un. | Un. | Un. | Un. | |
青枯菌 Ralstonia solanacearum | Un. | Un. | Un. | Un. | |
野油菜黄单胞菌 Xanthomonas campestris pv. campestris | Un. | Un. | Un. | Un. | |
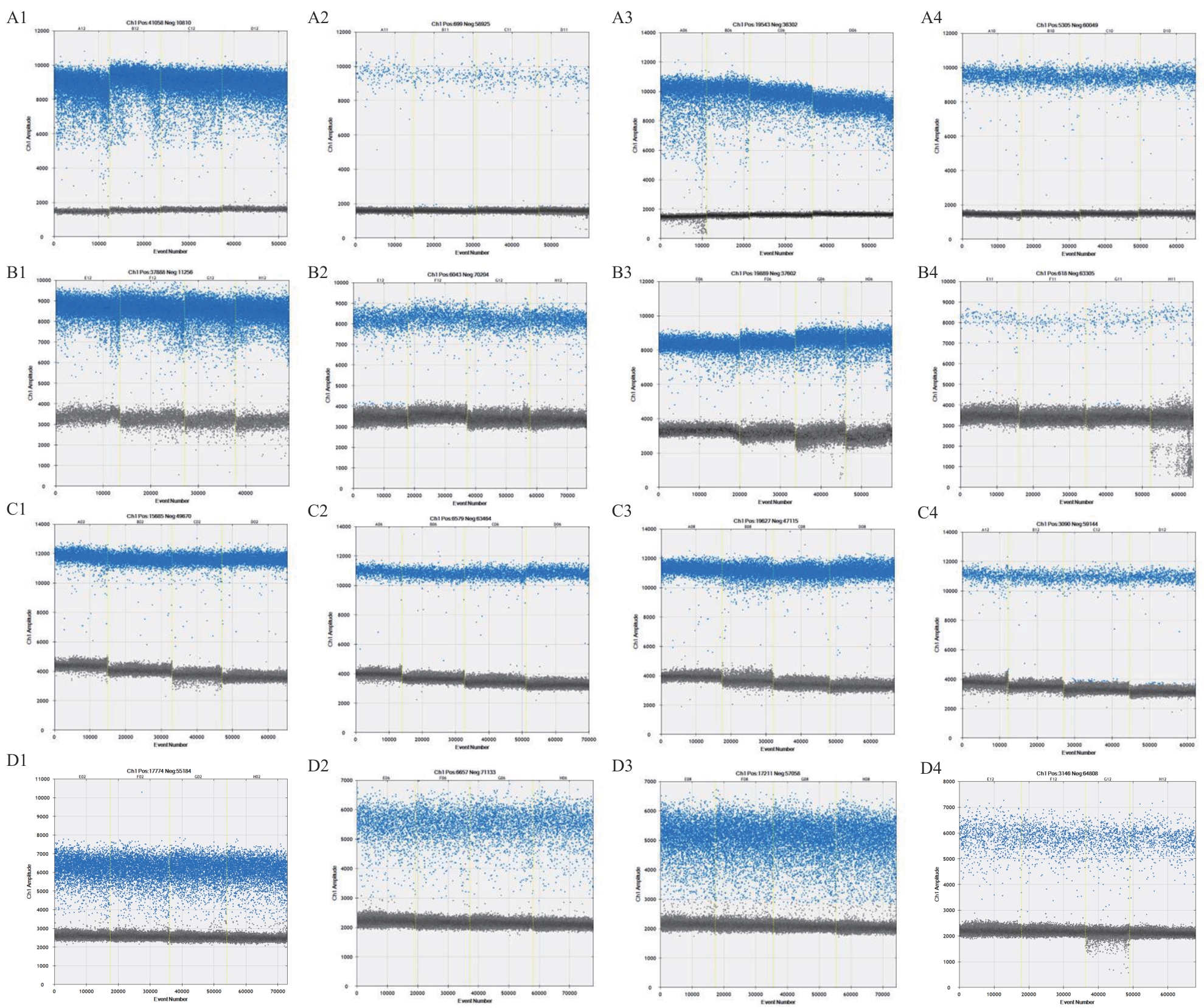
Fig. 3 ddPCR plots in different sets of primers and probesA-D: Primer combinations of ddPCR tests for Ver2, Ver3, Ver5 and Ver6, respectively. A1-D1: The raindrop map of Vd; A2-D2: the raindrop map of Vl; A3-D3: the raindrop pattern of Vna; A4-D4: the raindrop pattern of Vaa

Fig. 4 ddPCR results of Vd (A), Vl (B), Vna (C) and Vaa (D) at different annealing temperaturesThe 10 ddPCR reactions are divided by yellow dotted lines, and the annealing temperature gradient is 54-62 ℃. The blue is the positive drop, and gray is the negative drop

Fig. 5 ddPCR results with different concentration of primers and probe in Vd (A), Vl (B), Vna (C), and Vaa (D)Six ddPCR reactions are divided by vertical dotted yellow lines with a primer gradient ranged from 400-600 nmol/L, and a probe gradient ranged from100-400 nmol/L. The blue is the positive drop, and gray is the negative drop

Fig. 7 Test results of ddPCR sensitivityThe Vd (A), Vl (B), Vna (C) and Vaa (D) genomic DNAs were used as template. Seventy-six ddPCR reactions are divided by vertical dotted yellow lines with bacteria solution concentration from 2.1 × 10-3 ng/μL to 2.1 × 10-7 ng/μL, 1.6 × 10-3 ng/μL to 1.6 × 10-6 ng/μL, 6.9 × 10-2 ng/μL to 6.9 × 10-6 ng/μL and 3.6 × 10-3 ng/μL to 3.6 × 10-7 ng/μL, respectively. The pink line is the threshold, blue part is positive droplet and gray part is negative droplet
样本 Sample | 分析 Analysis | 阳性/全部(阳性率%) Positive/total(% Positive) |
|---|---|---|
植物 Plant | qPCR | 15/50(30%) |
| ddPCR | 22/50(44%) | |
土壤 Soil | qPCR | 27/50(54%) |
| ddPCR | 41/50(82%) |
Table 4 Detection of 50 cotton and soil samples using qPCR and ddPCR
样本 Sample | 分析 Analysis | 阳性/全部(阳性率%) Positive/total(% Positive) |
|---|---|---|
植物 Plant | qPCR | 15/50(30%) |
| ddPCR | 22/50(44%) | |
土壤 Soil | qPCR | 27/50(54%) |
| ddPCR | 41/50(82%) |
| [1] | Klosterman SJ, Subbarao KV, Kang S, et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens [J]. PLoS Pathog, 2011, 7(7): e1002137. |
| [2] | Pegg G F, Brady B L. Verticillium wilts [M]. New York: CABI Publishing, 2002. |
| [3] | Depotter JRL, Deketelaere S, Inderbitzin P, et al. Verticillium longisporum, the invisible threat to oilseed rape and other brassicaceous plant hosts [J]. Mol Plant Pathol, 2016, 17(7): 1004-1016. |
| [4] | Flajšman M, Radišek S, Javornik B. Pathogenicity assay of Verticillium nonalfalfae on hop plants [J]. Bio Protoc, 2017, 7(6): e2171. |
| [5] | Larsen RC, Vandemark GJ, Hughes TJ, et al. Development of a real-time polymerase chain reaction assay for quantifying Verticillium albo-atrum DNA in resistant and susceptible alfalfa [J]. Phytopathology, 2007, 97(11): 1519-1525. |
| [6] | Duressa D, Rauscher G, Koike ST, et al. A real-time PCR assay for detection and quantification of Verticillium dahliae in spinach seed [J]. Phytopathology, 2012, 102(4): 443-451. |
| [7] | Floerl S, Druebert C, Majcherczyk A, et al. Defence reactions in the apoplastic proteome of oilseed rape (Brassica napus var. napus) attenuate Verticillium longisporum growth but not disease symptoms [J]. BMC Plant Biol, 2008, 8: 129. |
| [8] | Wang D, Chen JY, Song J, et al. Cytotoxic function of xylanase VdXyn4 in the plant vascular wilt pathogen Verticillium dahliae [J]. Plant Physiol, 2021, 187(1): 409-429. |
| [9] | 索南措, 黄远志, 李彦忠, 等. 苜蓿黄萎病的发生、危害及检测 [J]. 草业科学, 2019, 36(9): 2384-2394. |
| Suo NC, Huang YZ, Li YZ, et al. Verticillium wilt of alfalfa: Occurrence, perniciousness, and detection[J]. Pratacultural Science, 2019, 36(9): 2384-2394 . | |
| [10] | Radišek S, Jakše J, Javornik B. Development of pathotype-specific SCAR markers for detection of Verticillium albo-atrum isolates from hop [J]. Plant Dis, 2004, 88(10): 1115-1122. |
| [11] | Griffiths DA. The development of lignitubers in roots after infection by Verticillium dahliae Kleb [J]. Can J Microbiol, 1971, 17(4): 441-444. |
| [12] | Isaac I. Verticillium wilt of sainfoin [J]. Ann Appl Biol, 1946, 33(1): 28-34. |
| [13] | Mercado-Blanco J, Rodríguez-Jurado D, Pérez-Artés E, et al. Detection of the nondefoliating pathotype of Verticillium dahliae in infected olive plants by nested PCR [J]. Plant Pathol, 2001, 50(5): 609-619. |
| [14] | Huang HC. Verticillium wilt of alfalfa: epidemiology and control strategies [J]. Can J Plant Pathol, 2003, 25(4): 328-338. |
| [15] | Bian XJ, Jing FX, Li G, et al. A microfluidic droplet digital PCR for simultaneous detection of pathogenic Escherichia coli O157 and Listeria monocytogenes [J]. Biosens Bioelectron, 2015, 74: 770-777. |
| [16] | Moukhamedov R. Use of polymerase chain reaction-amplified ribosomal intergenic sequences for the diagnosis of Verticillium tricorpus [J]. Phytopathology, 1994, 84(3): 256. |
| [17] | 朱荷琴, 李志芳, 冯自力, 等. 我国棉花黄萎病研究十年回顾及展望 [J]. 棉花学报, 2017, 29(S1): 37-50. |
| Zhu HQ, Li ZF, Feng ZL, et al. Overview of cotton Verticillium wilt research over the past decade in China and its prospect in future [J]. Cotton Science, 2017, 29(S1): 37-50. | |
| [18] | Gayoso C, de Ilárduya OM, Pomar F, et al. Assessment of real-time PCR as a method for determining the presence of Verticillium dahliae in different Solanaceae cultivars [J]. Eur J Plant Pathol, 2007, 118(3): 199-209. |
| [19] | Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments [J]. Clin Chem, 2009, 55(4): 611-622. |
| [20] | Mai GQ, Chen JY, Zhang M, et al. Construction of a pathogenic microorganism detection method based on third-generation nanopore sequencing data [J]. BMC Infect Dis, 2025, 25(1): 189. |
| [21] | Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool [J]. Clin Chem, 2015, 61(1): 79-88. |
| [22] | Sanders R, Huggett JF, Bushell CA, et al. Evaluation of digital PCR for absolute DNA quantification [J]. Anal Chem, 2011, 83(17): 6474-6484. |
| [23] | Alikian M, Whale AS, Akiki S, et al. RT-qPCR and RT-digital PCR: a comparison of different platforms for the evaluation of residual disease in chronic myeloid leukemia [J]. Clin Chem, 2017, 63(2): 525-531. |
| [24] | Pinheiro LB, Coleman VA, Hindson CM, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification [J]. Anal Chem, 2012, 84(2): 1003-1011. |
| [25] | Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR [J]. Nat Methods, 2013, 10(10): 1003-1005. |
| [26] | Wang D, Liu EL, Liu HY, et al. A droplet digital PCR assay for detection and quantification of Verticillium nonalfalfae and V. albo-atrum [J]. Front Cell Infect Microbiol, 2023, 12: 1110684. |
| [27] | Wang D, Jiao XY, Jia HJ, et al. Detection and quantification of Verticillium dahliae and V. longisporum by droplet digital PCR versus quantitative real-time PCR [J]. Front Cell Infect Microbiol, 2022, 12: 995705. |
| [28] | Moradi A, Almasi MA, Jafary H, et al. A novel and rapid loop-mediated isothermal amplification assay for the specific detection of Verticillium dahliae [J]. J Appl Microbiol, 2014, 116(4): 942-954. |
| [29] | Boogaerts T, Jacobs L, De Roeck N, et al. An alternative approach for bioanalytical assay optimization for wastewater-based epidemiology of SARS-CoV-2 [J]. Sci Total Environ, 2021, 789: 148043. |
| [30] | Ahmad NA, Heng LY, Salam F, et al. A colorimetric pH sensor based on Clitoria sp and Brassica sp for monitoring of food spoilage using chromametry [J]. Sensors, 2019, 19(21): 4813. |
| [31] | Dingle TC, Sedlak RH, Cook L, et al. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances [J]. Clin Chem, 2013, 59(11): 1670-1672. |
| [32] | 黄瑾, 梁涛波, 许恒毅. 数字PCR在生物学检测中应用的研究进展 [J]. 生命科学, 2021, 33(2): 255-264. |
| Huang J, Liang TB, Xu HY. Research progress of application of digital PCR in biological detection [J]. Chinese Bulletin of Life Sciences, 2021, 33(2): 255-264. | |
| [33] | Wang M, Yang JJ, Gai ZT, et al. Comparison between digital PCR and real-time PCR in detection of Salmonella typhimurium in milk [J]. Int J Food Microbiol, 2018, 266: 251-256. |
| [1] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [2] | ZHANG Yong, SONG Sheng-long, LI Yong-tai, ZHANG Xin-yu, LI Yan-jun. Cloning of GhSWEET9 in Upland Cotton and Functional Analysis of Resistance to Verticillium Wilt [J]. Biotechnology Bulletin, 2025, 41(6): 144-154. |
| [3] | SONG Hui-yang, SU Bao-jie, LI Jing-hao, MEI Chao, SONG Qian-na, CUI Fu-zhu, FENG Rui-yun. Cloning and Functional Analysis of the StAS2-15 Gene in Potato under Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 119-128. |
| [4] | DU Pin-ting, WU Guo-jiang, WANG Zhen-guo, LI Yan, ZHOU Wei, ZHOU Ya-xing. Identification and Expression Analysis of CPP Gene Family in Sorghum [J]. Biotechnology Bulletin, 2025, 41(1): 132-142. |
| [5] | LI Yong-hui, BAO Xing-xing, DUAN Yi-ke, ZHAO Yun-xia, YU Xiang-li, CHEN Yao, ZHANG Yan-zhao. Genome-wide Identification and Expression Features Analysis of the bZIP Family in Rhododendron henanense subsp. lingbaoense [J]. Biotechnology Bulletin, 2024, 40(8): 186-198. |
| [6] | ZHOU Jiang-hong, XIA Fei, ZHONG Li, QIU Lan-fen, LI Guang, LIU Qian, ZHANG Guo-feng, SHAO Jin-li, LI Na, CHE Shao-chen. Whole Genome Sequencing and Comparative Genomic Analysis of Antagonistic Bacterium CCBC3-3-1 against Verticillium dahlia [J]. Biotechnology Bulletin, 2024, 40(7): 235-246. |
| [7] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [8] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [9] | TIAN Shan-shan, HUANG Shi-yu, YANG Tian-wei, GAO Man-rong, ZHANG Shang-wen, HE Long-fei, ZHANG Xiang-jun, LI Ting, SHI Qian. Identification and Expression Analysis of MYB Gene Family in Dendrobium catenatum Under the Combined Stress of High Temperature and Drought [J]. Biotechnology Bulletin, 2024, 40(12): 145-159. |
| [10] | PAN Guo-qiang, WU Si-yuan, LIU Lu, GUO Hui-ming, CHENG Hong-mei, SU Xiao-feng. Construction and Preliminary Analysis of Verticillim dahliae Mutant Library [J]. Biotechnology Bulletin, 2023, 39(5): 112-119. |
| [11] | MU De-tian, WAN Ling-yun, ZHANG Yao, WEI Shu-gen, LU Ying, FU Jin-e, TIAN Yi, PAN Li-mei, TANG Qi. House-keeping Genes Screening and Expression Patterns Analysis of Genes Involved in Alkaloid Biosynthesis in Uncaria rhynchophylla [J]. Biotechnology Bulletin, 2023, 39(2): 126-138. |
| [12] | TENG Meng-xin, XU Ya, HE Jing, WANG Qi, QIAO Fei, LI Jing-yang, LI Xin-guo. Cloning and Prokaryotic Expression Analysis of MaMC6 in Banana [J]. Biotechnology Bulletin, 2023, 39(12): 179-186. |
| [13] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [14] | JIANG Xu-dong, LIU Yu, WU Jian-fei, HU Shuang-ge, LU Jian-yuan, ZI Xiang-dong. Tissue Expression and Localization Anaysis of FGG in Female Reproductive Organs of Bos grunniens [J]. Biotechnology Bulletin, 2022, 38(11): 286-294. |
| [15] | TIAN Li, LI Jun-jiao, DAI Xiao-feng, ZHANG Dan-dan, CHEN Jie-yin. From Functional Genes to Biological Characteristics:The Molecular Basis of Pathogenicity in Verticillium dahliae [J]. Biotechnology Bulletin, 2022, 38(1): 51-69. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||