








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 345-356.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0082
GUAN Zhi-hao1( ), SHAN Zhi-yi2,3, XIONG He1, ZHAO Rui-xue1,4(
), SHAN Zhi-yi2,3, XIONG He1, ZHAO Rui-xue1,4( )
)
Received:2025-01-19
Online:2025-09-26
Published:2025-09-24
Contact:
ZHAO Rui-xue
E-mail:gzhzjk445@outlook.com;zhaoruixue@caas.cn
GUAN Zhi-hao, SHAN Zhi-yi, XIONG He, ZHAO Rui-xue. Computational Literature-based Knowledge Discovery for Soybean Coupling Traits[J]. Biotechnology Bulletin, 2025, 41(9): 345-356.
| 实体类型Entity type | 实体开始位置标注 Annotation of entity start position | 实体内部位置标注 Annotation of entity internal position |
|---|---|---|
| 基因 Gene | B-GEN | I-GEN |
| 蛋白质 Protein | B-PRO | I-PRO |
| 性状 Trait | B-TRA | I-TRA |
| 非实体 Non-entity | O | O |
Table 1 Labeling strategy of soybean trait knowledge entities
| 实体类型Entity type | 实体开始位置标注 Annotation of entity start position | 实体内部位置标注 Annotation of entity internal position |
|---|---|---|
| 基因 Gene | B-GEN | I-GEN |
| 蛋白质 Protein | B-PRO | I-PRO |
| 性状 Trait | B-TRA | I-TRA |
| 非实体 Non-entity | O | O |
| 关系类型 Relation type | 关系标注 Relation annotation |
|---|---|
| e1上调e2 | UP(e1, e2) |
| e1下调e2 | DP(e1, e2) |
| e1与e2相关 | AW(e1, e2) |
Table 2 Relationship annotation strategy for soybean trait knowledge entities
| 关系类型 Relation type | 关系标注 Relation annotation |
|---|---|
| e1上调e2 | UP(e1, e2) |
| e1下调e2 | DP(e1, e2) |
| e1与e2相关 | AW(e1, e2) |
| 文本序列 Text sequence | 文本标注 Text annotation |
|---|---|
| A | O |
| GmPP2C-1 | B-GEN |
| Gene | O |
| From | O |
| Wild | O |
| Soybean | O |
| Helps | O |
| To | O |
| Increase | O |
| Seed | B-TRA |
| Weight | I-TRA |
| UP (GmPP2C-1, seed weight) |
Table 3 Examples of text annotation
| 文本序列 Text sequence | 文本标注 Text annotation |
|---|---|
| A | O |
| GmPP2C-1 | B-GEN |
| Gene | O |
| From | O |
| Wild | O |
| Soybean | O |
| Helps | O |
| To | O |
| Increase | O |
| Seed | B-TRA |
| Weight | I-TRA |
| UP (GmPP2C-1, seed weight) |
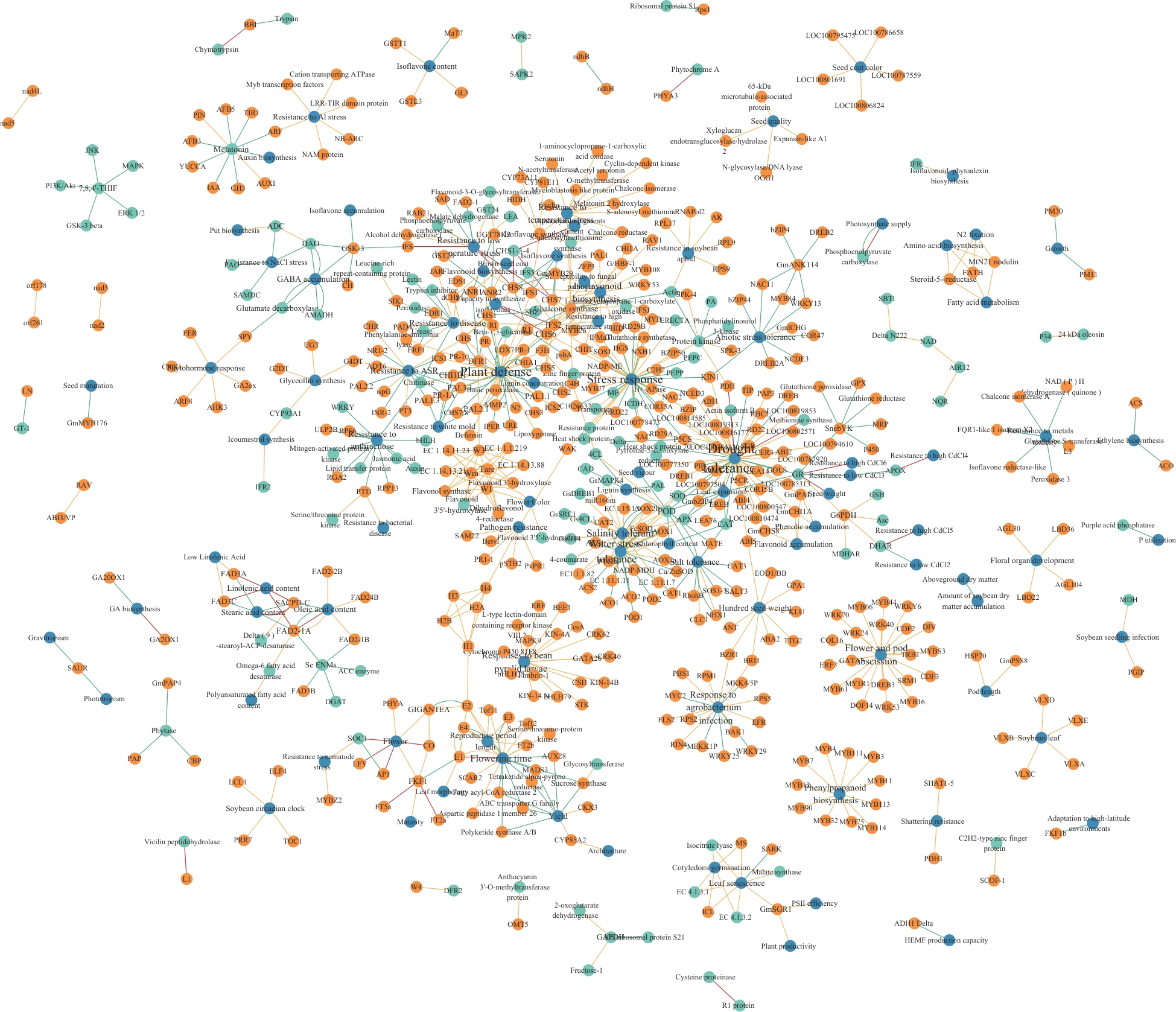
Fig. 2 Knowledge network of soybean traitOrange nodes indicate gene entities, green nodes indicate protein entities, blue nodes indicate trait entities, green edges indicate "upregulate" relationships, red edges indicate "downregulate" relationships, and yellow edges indicate "associated with" relationships. The same below
| 网络属性 Network property | 数值 Value |
|---|---|
| 节点数量 Number of nodes | 648 |
| 连边数量 Number of edges | 774 |
| 平均邻居数 Average number of neighbors | 2.511 |
| 网络直径 Diameter of network | 19 |
| 网络半径 Radius of network | 10 |
| 特征路径长度 Characteristic path length | 6.934 |
| 聚类系数 Coefficient of clustering | 0.034 |
| 网络密度 Density of network | 0.006 |
| 网络异质性 Network heterogeneity | 1.780 |
| 网络中心化 Centralization of the network | 0.126 |
| 连通分量 Component of connectivity | 50 |
Table 4 Topological properties of the network
| 网络属性 Network property | 数值 Value |
|---|---|
| 节点数量 Number of nodes | 648 |
| 连边数量 Number of edges | 774 |
| 平均邻居数 Average number of neighbors | 2.511 |
| 网络直径 Diameter of network | 19 |
| 网络半径 Radius of network | 10 |
| 特征路径长度 Characteristic path length | 6.934 |
| 聚类系数 Coefficient of clustering | 0.034 |
| 网络密度 Density of network | 0.006 |
| 网络异质性 Network heterogeneity | 1.780 |
| 网络中心化 Centralization of the network | 0.126 |
| 连通分量 Component of connectivity | 50 |

Fig. 3 Connected subnetworks s of gravitropism and phototropism (A), seed composition control (B), stay green (C), oil quality (D) and resistance to Al stress (E) coupling traits except for maximally connected subnetwork
| 性状1 Trait 1 | 路径 Path | 性状2 Trait 2 | WOS入藏号 WOS accession number | 说明 Explanation |
|---|---|---|---|---|
| Drought tolerance | KIN1 | Abiotic stress tolerance | 001021178600001 | 耐旱和耐非生物胁迫性状受KIN1调节 |
| CAT | Chlorophyll content | 001270505200012 | 耐旱和叶绿素含量性状受CAT和POD调节 | |
| POD | 001332116100001 | |||
| GmCHI1A | Flavonoid accumulation | 001095996200006 | 耐旱和类黄酮累积性状受GmCHI1A、GmCHS8和GmPAL1调节 | |
| GmCHS8 | ||||
| GmPAL1 | ||||
| ABI5 | Hundred seed weight | 001290873600001 | 耐旱和百粒重性状受ABI5调节 | |
| APX | Leaf expansion | 001094244600002 | 耐旱和叶片扩张性状受APX、CAT和SOD调节 | |
| CAT | ||||
| SOD | 000934257600001 | |||
| POD | Lignin synthesis | 001069057100001 | 耐旱和木质素合成性状受POD调节 | |
| GmCHI1A | Phenolic accumulation | 001095996200006 | 耐旱和酚累积性状受GmCHI1A、GmCHS8和GmPAL1调节 | |
| GmCHS8 | ||||
| GmPAL1 | ||||
| GR | Resistance to high CdCl6 | 001093350900063 | 耐旱和耐铬胁迫性状受GR调节 | |
| GR | Resistance to low CdCl3 | |||
| CAT | Salt tolerance | 000950283400001 | 耐旱和耐盐性状受CAT和POD调节 | |
| POD | 001072139900001 | |||
| POD | Seed vigour | 000896181600001 | 耐旱和种子活力性状受POD调节 | |
| GR | Seed weight | 000689953000001 | 耐旱和种子重量性状受GR调节 | |
| BZIP | Stress response | 000859460000001 | 耐旱和耐胁迫性状受BZIP和P5CS调节 | |
| P5CS | 000253050100012 |
Table 5 Traits coupling to drought tolerance
| 性状1 Trait 1 | 路径 Path | 性状2 Trait 2 | WOS入藏号 WOS accession number | 说明 Explanation |
|---|---|---|---|---|
| Drought tolerance | KIN1 | Abiotic stress tolerance | 001021178600001 | 耐旱和耐非生物胁迫性状受KIN1调节 |
| CAT | Chlorophyll content | 001270505200012 | 耐旱和叶绿素含量性状受CAT和POD调节 | |
| POD | 001332116100001 | |||
| GmCHI1A | Flavonoid accumulation | 001095996200006 | 耐旱和类黄酮累积性状受GmCHI1A、GmCHS8和GmPAL1调节 | |
| GmCHS8 | ||||
| GmPAL1 | ||||
| ABI5 | Hundred seed weight | 001290873600001 | 耐旱和百粒重性状受ABI5调节 | |
| APX | Leaf expansion | 001094244600002 | 耐旱和叶片扩张性状受APX、CAT和SOD调节 | |
| CAT | ||||
| SOD | 000934257600001 | |||
| POD | Lignin synthesis | 001069057100001 | 耐旱和木质素合成性状受POD调节 | |
| GmCHI1A | Phenolic accumulation | 001095996200006 | 耐旱和酚累积性状受GmCHI1A、GmCHS8和GmPAL1调节 | |
| GmCHS8 | ||||
| GmPAL1 | ||||
| GR | Resistance to high CdCl6 | 001093350900063 | 耐旱和耐铬胁迫性状受GR调节 | |
| GR | Resistance to low CdCl3 | |||
| CAT | Salt tolerance | 000950283400001 | 耐旱和耐盐性状受CAT和POD调节 | |
| POD | 001072139900001 | |||
| POD | Seed vigour | 000896181600001 | 耐旱和种子活力性状受POD调节 | |
| GR | Seed weight | 000689953000001 | 耐旱和种子重量性状受GR调节 | |
| BZIP | Stress response | 000859460000001 | 耐旱和耐胁迫性状受BZIP和P5CS调节 | |
| P5CS | 000253050100012 |
性状1 Trait 1 | 路径 Path | 性状2 Trait 2 | WOS入藏号 Accession number | 说明 Explanation |
|---|---|---|---|---|
| Chlorophyll content | POD | Drought tolerance | 001329603400001 | 叶绿素含量与耐旱、叶片扩张和木质素合成性状耦合 |
| POD | Leaf expansion | A1997XJ61200003 | ||
| POD | Lignin synthesis | 000825131700001 | ||
| Lignin synthesis | POD | Drought tolerance | 000619359600001 | 木质素合成与耐旱、叶片扩张、耐盐和耐水胁迫性状耦合 |
| POD | Leaf expansion | 000372112300005 | ||
| POD | Salinity tolerant | 001294925800001 | ||
| POD | Water stress tolerance | 000368074100001 | ||
| Salinity tolerant | POD | Chlorophyll content | 000465414300006 | 耐盐与叶绿素含量、耐旱、叶片扩张、木质素合成和种子活力性状耦合 |
| POD | Drought tolerance | 000738259400001 | ||
| POD | Leaf expansion | 000723907300001 | ||
| POD | Lignin synthesis | 001062757700002 | ||
| POD | Seed vigour | 000896181600001 | ||
| Seed vigour | POD | Drought tolerance | 000532138700001 | 种子活力与耐旱、叶片扩张、木质素合成和耐盐性状耦合 |
| POD | Leaf expansion | 000230397200029 | ||
| POD | Lignin synthesis | 001131146600001 | ||
| POD | Salinity tolerant | 000427900200004 |
Table 6 Coupling traits in POD pathways
性状1 Trait 1 | 路径 Path | 性状2 Trait 2 | WOS入藏号 Accession number | 说明 Explanation |
|---|---|---|---|---|
| Chlorophyll content | POD | Drought tolerance | 001329603400001 | 叶绿素含量与耐旱、叶片扩张和木质素合成性状耦合 |
| POD | Leaf expansion | A1997XJ61200003 | ||
| POD | Lignin synthesis | 000825131700001 | ||
| Lignin synthesis | POD | Drought tolerance | 000619359600001 | 木质素合成与耐旱、叶片扩张、耐盐和耐水胁迫性状耦合 |
| POD | Leaf expansion | 000372112300005 | ||
| POD | Salinity tolerant | 001294925800001 | ||
| POD | Water stress tolerance | 000368074100001 | ||
| Salinity tolerant | POD | Chlorophyll content | 000465414300006 | 耐盐与叶绿素含量、耐旱、叶片扩张、木质素合成和种子活力性状耦合 |
| POD | Drought tolerance | 000738259400001 | ||
| POD | Leaf expansion | 000723907300001 | ||
| POD | Lignin synthesis | 001062757700002 | ||
| POD | Seed vigour | 000896181600001 | ||
| Seed vigour | POD | Drought tolerance | 000532138700001 | 种子活力与耐旱、叶片扩张、木质素合成和耐盐性状耦合 |
| POD | Leaf expansion | 000230397200029 | ||
| POD | Lignin synthesis | 001131146600001 | ||
| POD | Salinity tolerant | 000427900200004 |
| [1] | Tam V, Patel N, Turcotte M, et al. Benefits and limitations of genome-wide association studies [J]. Nat Rev Genet, 2019, 20(8): 467-484. |
| [2] | Nédellec C, Sauvion C, Bossy R, et al. TaeC: a manually annotated text dataset for trait and phenotype extraction and entity linking in wheat breeding literature [J]. PLoS One, 2024, 19(6): e0305475. |
| [3] | Yacoubi Ayadi N, Bernard S, Bossy R, et al. A unified approach to publish semantic annotations of agricultural documents as knowledge graphs [J]. Smart Agric Technol, 2024, 8: 100484. |
| [4] | Gao YJ, Zhou Q, Luo JX, et al. Crop-GPA an integrated platform of crop gene-phenotype associations [J]. NPJ Syst Biol Appl, 2024, 10(1): 15. |
| [5] | Lotreck S, Segura Abá K, Lehti-Shiu MD, et al. Plant science knowledge graph corpus: a gold standard entity and relation corpus for the molecular plant sciences [J]. Silico Plants, 2024, 6(1): diad021. |
| [6] | Lee HJ, Chung YJ, Jang S, et al. Genome-wide identification of major genes and genomic prediction using high-density and text-mined gene-based SNP panels in Hanwoo (Korean cattle) [J]. PLoS One, 2020, 15(12): e0241848. |
| [7] | Singh G, Papoutsoglou EA, Keijts-Lalleman F, et al. Extracting knowledge networks from plant scientific literature: potato tuber flesh color as an exemplary trait [J]. BMC Plant Biol, 2021, 21(1): 198. |
| [8] | Xie CJ, Gao J, Chen JJ, et al. PotatoG-DKB a potato gene-disease knowledge base mined from biological literature [J]. PeerJ, 2024, 12: e18202. |
| [9] | Liu YY. DKG-PIPD: a novel method about building deep knowledge graph [J]. IEEE Access, 2021, 9: 137295-137308. |
| [10] | Lu J, Yang WX, He L, et al. A method for extracting fine-grained knowledge of the wheat production chain [J]. Agronomy, 2024, 14(9): 1903. |
| [11] | Yuan WW, Yang WX, He L, et al. Research on entity and relationship extraction with small training samples for cotton pests and diseases [J]. Agriculture, 2024, 14(3): 457. |
| [12] | Yang K, Liu Y. Construction of knowledge graph in the field of grassland plants based on ontology database[C]//2021 International Conference on Environmental Remote Sensing and Big Data. Wuhan China: SPIE, 2021: 12129. |
| [13] | Lou DJ, Li F, Ge JY, et al. LncPheDB: a genome-wide lncRNAs regulated phenotypes database in plants [J]. aBIOTECH, 2022, 3(3): 169-177. |
| [14] | Fang C, Ma YM, Wu SW, et al. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean [J]. Genome Biol, 2017, 18(1): 161. |
| [15] | Singh G, Kuzniar A, Brouwer M, et al. Linked data platform for Solanaceae species [J]. Appl Sci, 2020, 10(19): 6813. |
| [16] | Ivanisenko TV, Saik OV, Demenkov PS, et al. The Solanum tuberosum knowledge base: the section on molecular-genetic regulation of metabolic pathways [J]. Vestn VOGiS, 2018, 22(1): 8-17. |
| [17] | Brown AV, Conners SI, Huang W, et al. A new decade and new data at SoyBase, the USDA-ARS soybean genetics and genomics database [J]. Nucleic Acids Res, 2021, 49(D1): D1496-D1501. |
| [18] | Zhang F, Ma LD, Wang JP, et al. An MRC and adaptive positive-unlabeled learning framework for incompletely labeled named entity recognition [J]. Int J Intell Syst, 2022, 37(11): 9580-9597. |
| [19] | Wu SC, He YF. Enriching pre-trained language model with entity information for relation classification [C]//Proceedings of the 28th ACM International Conference on Information and Knowledge Management. Beijing China. New York: ACM, 2019: 2361-2364. |
| [20] | Yuan XB, Jiang XY, Zhang MZ, et al. Integrative omics analysis elucidates the genetic basis underlying seed weight and oil content in soybean [J]. Plant Cell, 2024, 36(6): 2160-2175. |
| [21] | Wang XY, Yu RB, Wang JJ, et al. The asymmetric expression of SAUR genes mediated by ARF7/19 promotes the gravitropism and phototropism of plant hypocotyls [J]. Cell Rep, 2020, 31(3): 107529. |
| [22] | Wang CX, Ji YH, Cao XS, et al. Carbon dots improve nitrogen bioavailability to promote the growth and nutritional quality of soybeans under drought stress [J]. ACS Nano, 2022, 16(8): 12415-12424. |
| [23] | Shi SY, Miao HY, Du XM, et al. GmSGR1, a stay-green gene in soybean (Glycine max L.), plays an important role in regulating early leaf-yellowing phenotype and plant productivity under nitrogen deprivation [J]. Acta Physiol Plant, 2016, 38(4): 97. |
| [24] | Zhang JP, Wang XZ, Lu YM, et al. Genome-wide scan for seed composition provides insights into soybean quality improvement and the impacts of domestication and breeding [J]. Mol Plant, 2018, 11(3): 460-472. |
| [25] | Hassani-Pak K, Singh A, Brandizi M, et al. KnetMiner: a comprehensive approach for supporting evidence-based gene discovery and complex trait analysis across species [J]. Plant Biotechnol J, 2021, 19(8): 1670-1678. |
| [1] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [2] | LIAN Shao-jie, TANG Sheng-shuo, KANG Chuan-li, LIU Lei, ZHENG De-qiang, DU Shuai, TANG Li-wei, ZHANG Mei-xia, LIU Qiang. Isolation, Identification, Optimization of Fermentation Conditions of High-yield Tremella fuciformis Polysaccharides Enzyme-producing Strain and Its Enzyme Characteristics Analysis [J]. Biotechnology Bulletin, 2025, 41(9): 302-313. |
| [3] | CAO Yuan-yuan, ZHOU Shu-hao, ZHANG Hai-rong, CUI Xiao-na. Unfolded Protein Response Enhances Plant Resistance to Disease by Regulating Tryptophan Metabolism [J]. Biotechnology Bulletin, 2025, 41(8): 155-164. |
| [4] | LI Ya, JIANG Lin, XU Chuang, WANG Su-hui, MA Zhao, WANG Liang. Research Progress in Molecular Defense Mechanisms of Chlamydomonas reinhardtii in Response to Heavy Metal Stress [J]. Biotechnology Bulletin, 2025, 41(8): 53-64. |
| [5] | LI De-hai, YIN Li, ZHOU Cai-xue, WANG Ze-tong, SUN Chang-yan. Screening, Identification and Preservation of Lactic Acid Bacteria Inhibiting the Growth of Rot-causing Fungus Lonicera caerulea L. [J]. Biotechnology Bulletin, 2025, 41(8): 289-299. |
| [6] | ZHANG Xue-qiong, PAN Su-jun, LI Wei, DAI Liang-ying. Research Progress of Plant Phosphate Transporters in the Response to Stress [J]. Biotechnology Bulletin, 2025, 41(7): 28-36. |
| [7] | GAO Jing, CHENG Yi-cun, GAO Ming, ZHAO Yun-xiao, WANG Yang-dong. Regulation of Plant Tannin Synthesis and Mechanisms of Its Responses to Environment [J]. Biotechnology Bulletin, 2025, 41(7): 49-59. |
| [8] | LI Si-bo, QIAN Hong-ping, XU Chang-wen, WANG Xiao, LIN Jin-xing, CUI Ya-ning. Research Progress in the Involvement of Intracellular Transport Regulated by Endogenous Elicitors in Plant Growth and Development and Response to Adverse Stress [J]. Biotechnology Bulletin, 2025, 41(7): 17-27. |
| [9] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [10] | LUO Ji-lin, LI Jin-ye, JIA Yu-xin. Identification and Functional Analysis of Gravity Response Regulatory Genes in Potato [J]. Biotechnology Bulletin, 2025, 41(6): 109-118. |
| [11] | LI Rui, HU Ting, CHEN Shu-wei, WANG Yao, WANG Ji-ping. Positive Regulation of Anthocyanin Biosynthesis by PfMYB80 Transcription Factor in Perilla frutescens [J]. Biotechnology Bulletin, 2025, 41(6): 243-255. |
| [12] | WU Hao, DONG Wei-feng, HE Zi-tian, LI Yan-xiao, XIE Hui, SUN Ming-zhe, SHEN Yang, SUN Xiao-li. Genome-wide Identification and Expression Analysis of the Rice BXL Gene Family [J]. Biotechnology Bulletin, 2025, 41(6): 87-98. |
| [13] | ZHANG Ji-chang, XU Yun-feng, JIANG Ling-yan. Optimization of Fermentation Conditions of Endophytic Bacterium ZW21 Isolated from Stylosanthes and Stability Analysis of Antimicrobial Substances [J]. Biotechnology Bulletin, 2025, 41(5): 280-289. |
| [14] | LIU Zhuo-jun, CHAI Wen-ting, REN Yi-le, WANG Xin-yu, ZHU Li-xun, ZHAO Shan-shan, YANG Bo-hui, FAN Jia-li, LI Xin-feng, ZHAO Wei-jun, LYU Jin-hui, ZHANG Chun-lai. Analysis on Expression and DNA Variation of TGA Genes in Sorghum (Sorghum bicolor) in Response to Sporisorium reilianum Infection [J]. Biotechnology Bulletin, 2025, 41(5): 90-103. |
| [15] | FAN Yue-ni, XIAN Bao-shan, SHI Yi-ping, REN Meng-yuan, XU Jia-hui, WEI Shao-wei, XU Xiao-jing, LUO Xiao-feng, SHU Kai. SPINDLY and SECRET AGENT-mediated Protein Glycosylation Regulates Plant Development and Stress Response [J]. Biotechnology Bulletin, 2025, 41(4): 1-8. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||