








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (10): 264-276.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0353
Previous Articles Next Articles
HAN Yu1( ), YUAN Qing-yun1,2, ZHANG Qing-ping1, WU Chun-lai1, HE Wei1(
), YUAN Qing-yun1,2, ZHANG Qing-ping1, WU Chun-lai1, HE Wei1( ), ZHANG Fen1(
), ZHANG Fen1( )
)
Received:2025-04-02
Online:2025-10-26
Published:2025-10-28
Contact:
HE Wei, ZHANG Fen
E-mail:2311914325@qq.com;guiliuer@163.com;zhangfen2008.cool@163.com
HAN Yu, YUAN Qing-yun, ZHANG Qing-ping, WU Chun-lai, HE Wei, ZHANG Fen. Cloning and Expression Analysis of Six Genes of the NPF Family in Tea Plants and Functional Verification of CsNPF7.3[J]. Biotechnology Bulletin, 2025, 41(10): 264-276.
目的 Purpose | 引物名称 Primer name | 前引物序列 Forward primer sequence (5′-3′) | 后引物序列 Reverse primer sequence (5′-3′) |
|---|---|---|---|
基因克隆 Gene cloneing | CsNPF2.13 | CCCAAGATAAGGAAAAATGGAGGTA | CATAGATTTGTCAAAGTGACTTCAT |
| CsNPF2.7 | AGACTCACACTCGGAGGCATAAACT | ACAAACAACACCACAACAACAGATG | |
| CsNPF3.1 | ATCCGACCGCCTCTACTCT | AGTTACAAACATATCGTTTAAGCTA | |
| CsNPF5.5 | GTGAGAGTGGAAAAGACAACGCATA | CCTAACAGCAGCAGCAACCCT | |
| CsNPF7.1 | TGGGGCTGTTTTATTAGTTGAG | AATATAGCCCGAAATAAGCCTGCA | |
| CsNPF7.3 | CCCAAACACCAACTTCTCTTCTCTC | CCTCACACAAATGCCTACTGTCCTT | |
表达分析 RT-qPCR analysis | CsGAPDH | TTGGCATCGTTGAGGGTCT | CAGTGGGAACACGGAAAGC |
| CsNPF2.13-q | AGGAAGGAAAGGAATCAACAGC | AACCTCGTCCCAACAAAGAAC | |
| CsNPF2.7-q | CGAACGCTGACACCTCTACA | GAGCCGCTTTAACTCCACCA | |
| CsNPF3.1-q | GCCACCTAATCCGAGGGATG | TGCCCAACTCAAGTGGACTG | |
| CsNPF5.5-q | GGCTAACTTTCATCGCCTGC | CAAGCTTGCGGTTCATGTGG | |
| CsNPF6.1-q | CGATAGCAGCGTGTTTGTGG | GGCTGCCTTGTCCAAACATC | |
| CsNPF6.3-q | GCCTCACTGACTGTTTTCTTCG | AACCGCTTGATTTCGCCTA | |
| CsNPF7.1-q | TAGCCACATTCGGAGCAGAC | CCGACGTTGAGTGCAGAGTA | |
| CsNPF7.3-q | GCCAACCGGCAATACGAGAA | GACATTATTAGCTGCGTCGGC | |
| Atβ-tubulin-q | ACCACTCCTAGCTTTGGTGATCTG | AGGTTCACTGCGAGCTTCCTCA |
Table 1 Primers’ sequences for gene cloning and RT-qPCR analysis
目的 Purpose | 引物名称 Primer name | 前引物序列 Forward primer sequence (5′-3′) | 后引物序列 Reverse primer sequence (5′-3′) |
|---|---|---|---|
基因克隆 Gene cloneing | CsNPF2.13 | CCCAAGATAAGGAAAAATGGAGGTA | CATAGATTTGTCAAAGTGACTTCAT |
| CsNPF2.7 | AGACTCACACTCGGAGGCATAAACT | ACAAACAACACCACAACAACAGATG | |
| CsNPF3.1 | ATCCGACCGCCTCTACTCT | AGTTACAAACATATCGTTTAAGCTA | |
| CsNPF5.5 | GTGAGAGTGGAAAAGACAACGCATA | CCTAACAGCAGCAGCAACCCT | |
| CsNPF7.1 | TGGGGCTGTTTTATTAGTTGAG | AATATAGCCCGAAATAAGCCTGCA | |
| CsNPF7.3 | CCCAAACACCAACTTCTCTTCTCTC | CCTCACACAAATGCCTACTGTCCTT | |
表达分析 RT-qPCR analysis | CsGAPDH | TTGGCATCGTTGAGGGTCT | CAGTGGGAACACGGAAAGC |
| CsNPF2.13-q | AGGAAGGAAAGGAATCAACAGC | AACCTCGTCCCAACAAAGAAC | |
| CsNPF2.7-q | CGAACGCTGACACCTCTACA | GAGCCGCTTTAACTCCACCA | |
| CsNPF3.1-q | GCCACCTAATCCGAGGGATG | TGCCCAACTCAAGTGGACTG | |
| CsNPF5.5-q | GGCTAACTTTCATCGCCTGC | CAAGCTTGCGGTTCATGTGG | |
| CsNPF6.1-q | CGATAGCAGCGTGTTTGTGG | GGCTGCCTTGTCCAAACATC | |
| CsNPF6.3-q | GCCTCACTGACTGTTTTCTTCG | AACCGCTTGATTTCGCCTA | |
| CsNPF7.1-q | TAGCCACATTCGGAGCAGAC | CCGACGTTGAGTGCAGAGTA | |
| CsNPF7.3-q | GCCAACCGGCAATACGAGAA | GACATTATTAGCTGCGTCGGC | |
| Atβ-tubulin-q | ACCACTCCTAGCTTTGGTGATCTG | AGGTTCACTGCGAGCTTCCTCA |
基因名称 Gene name | 开放阅读框长度 Open reading frame (ORF) (bp) | 编码氨基酸数目 Number of amino acids |
|---|---|---|
| CsNPF2.13 | 1 923 | 640 |
| CsNPF2.7 | 1 470 | 489 |
| CsNPF3.1 | 1 815 | 604 |
| CsNPF5.5 | 1 659 | 552 |
| CsNPF7.1 | 1 734 | 577 |
| CsNPF7.3 | 1 800 | 599 |
Table 2 Sequence information of CsNPFs in tea plants
基因名称 Gene name | 开放阅读框长度 Open reading frame (ORF) (bp) | 编码氨基酸数目 Number of amino acids |
|---|---|---|
| CsNPF2.13 | 1 923 | 640 |
| CsNPF2.7 | 1 470 | 489 |
| CsNPF3.1 | 1 815 | 604 |
| CsNPF5.5 | 1 659 | 552 |
| CsNPF7.1 | 1 734 | 577 |
| CsNPF7.3 | 1 800 | 599 |
蛋白 Protein | 蛋白分子量 Molecular weight (kD) | 蛋白等电点 Theoretical pI | 总原子数 Total number of atom | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 亲疏水性平均系数 Grand average of hydropathicity (GRAVY) | 信号肽 Signal peptide | 亚细胞定位 Sub-cellular localization | 跨膜螺旋预测 Transmembrane helices prediction |
|---|---|---|---|---|---|---|---|---|---|
| CsNPF2.13 | 70.26 | 8.82 | 9 950 | 46.14 | 97.66 | 0.228 | 0.001 3 | 质膜(8.48) | 12 |
| CsNPF2.7 | 53.62 | 8.81 | 7 633 | 42.13 | 108.4 | 0.332 | 0.287 5 | 质膜(9.4) | 12 |
| CsNPF3.1 | 67.10 | 6.11 | 9 523 | 32.75 | 104.59 | 0.183 | 0.000 5 | 质膜(8.3) | 11 |
| CsNPF5.5 | 61.38 | 8.70 | 8 713 | 32.29 | 102.46 | 0.375 | 0.526 5 | 质膜(8.5) | 9 |
| CsNPF7.1 | 62.92 | 6.05 | 8 849 | 25.63 | 97.14 | 0.337 | 0.004 6 | 质膜(8.4) | 12 |
| CsNPF7.3 | 66.51 | 8.53 | 9 378 | 29.64 | 96.51 | 0.263 | 0.002 0 | 质膜(9.1) | 12 |
Table 3 Basic characteristics of CsNPFs in tea plants
蛋白 Protein | 蛋白分子量 Molecular weight (kD) | 蛋白等电点 Theoretical pI | 总原子数 Total number of atom | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 亲疏水性平均系数 Grand average of hydropathicity (GRAVY) | 信号肽 Signal peptide | 亚细胞定位 Sub-cellular localization | 跨膜螺旋预测 Transmembrane helices prediction |
|---|---|---|---|---|---|---|---|---|---|
| CsNPF2.13 | 70.26 | 8.82 | 9 950 | 46.14 | 97.66 | 0.228 | 0.001 3 | 质膜(8.48) | 12 |
| CsNPF2.7 | 53.62 | 8.81 | 7 633 | 42.13 | 108.4 | 0.332 | 0.287 5 | 质膜(9.4) | 12 |
| CsNPF3.1 | 67.10 | 6.11 | 9 523 | 32.75 | 104.59 | 0.183 | 0.000 5 | 质膜(8.3) | 11 |
| CsNPF5.5 | 61.38 | 8.70 | 8 713 | 32.29 | 102.46 | 0.375 | 0.526 5 | 质膜(8.5) | 9 |
| CsNPF7.1 | 62.92 | 6.05 | 8 849 | 25.63 | 97.14 | 0.337 | 0.004 6 | 质膜(8.4) | 12 |
| CsNPF7.3 | 66.51 | 8.53 | 9 378 | 29.64 | 96.51 | 0.263 | 0.002 0 | 质膜(9.1) | 12 |
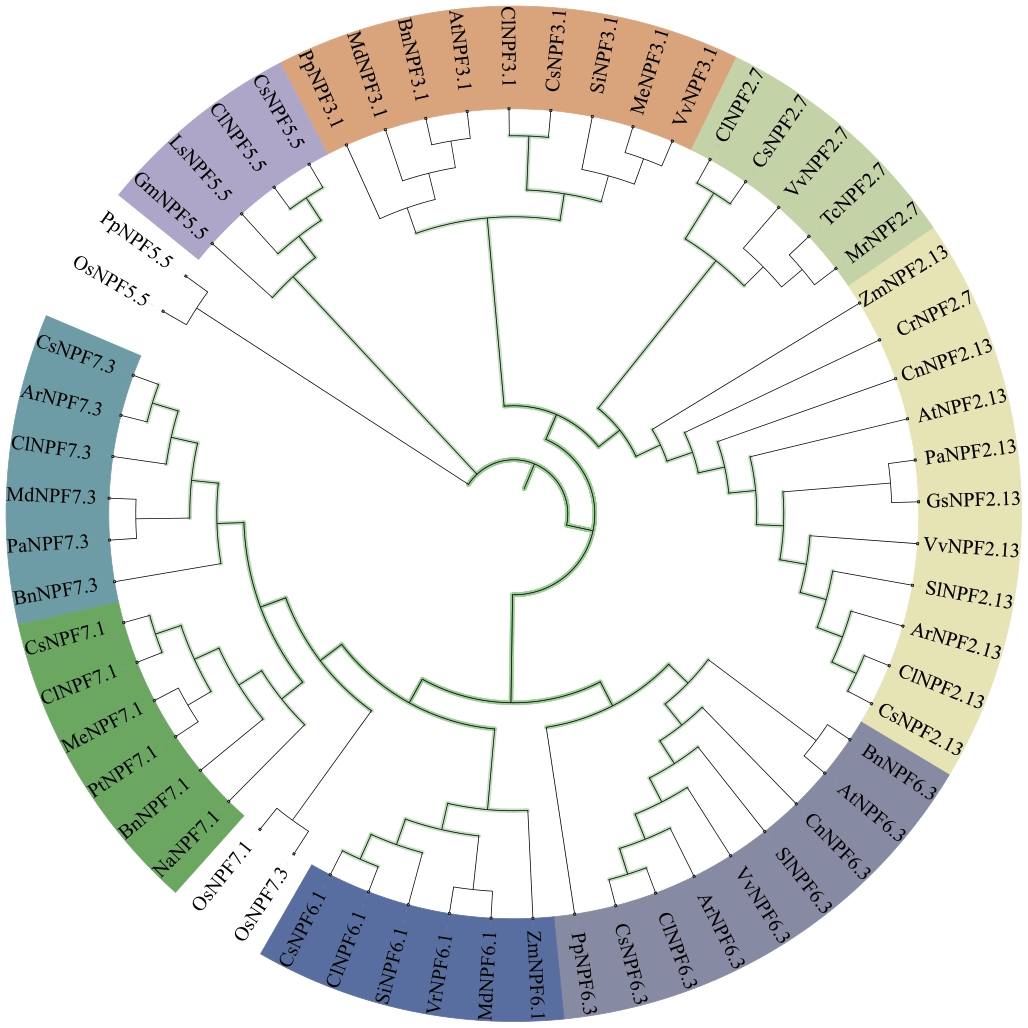
Fig. 2 Phylogenetic tree ofCsNPFs proteinsAt: Arabidopsis thaliana; Os: Oryza sativa; Zm: Zea mays; Mt: Medicago truncatula; Md: Malus domestica; Cl: Camellia lanceoleosa; Vv: Vitis vinifera; Si: Sesamum indicum; Pa: Populus alba; Cn: Cocos nucifera; Cr: Catharanthus roseus; Tc: Theobroma cacao; Pp: Pinus pinaster; Bn: Brassica napus; Gm: Glycine max; Na: Nicotiana attenuata;Pt: Populus trichocarpa; Ls: Lactuca sativais; Me: Manihot esculenta; Mr: Morella rubra; Ar: Actinidia rufa; Vr: Vitis riparis; Sl: Solanum lycopersicum; Gs: Glycine soja
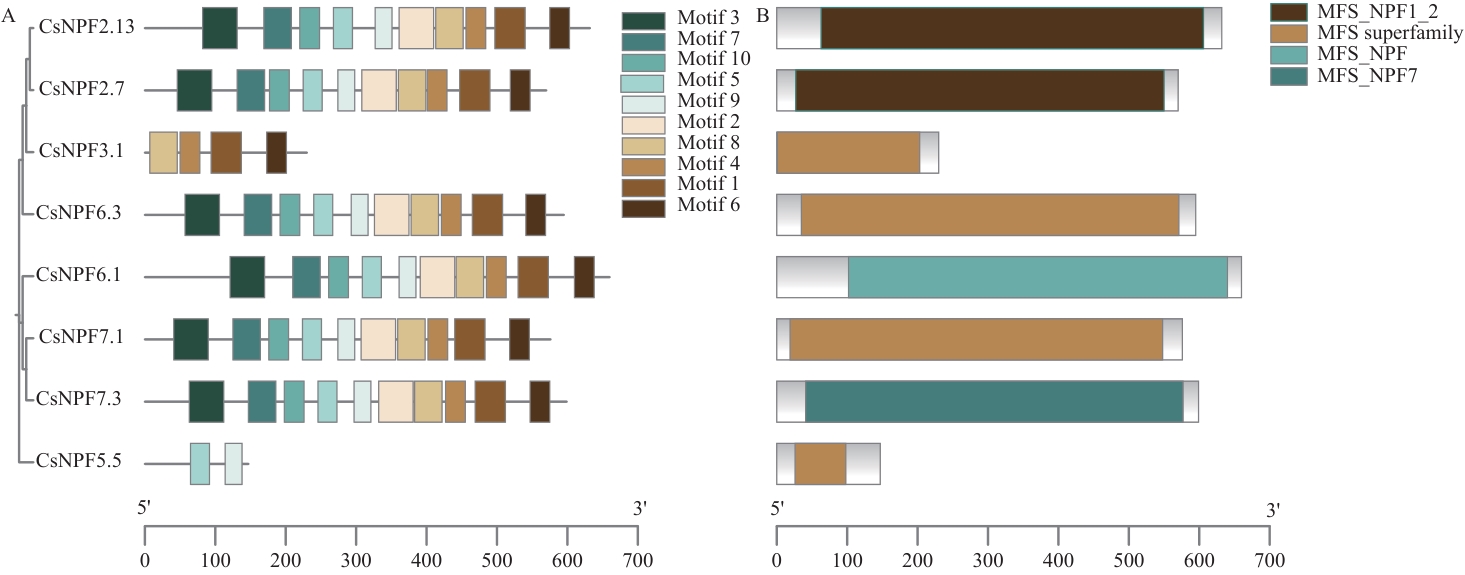
Fig. 3 Conserved motif (A) and domain (B) analysis of CsNPFs proteinsMFS_NPF1_2, MFS superfamily, and MFS_NPF7 denote nitrate transport-related domains
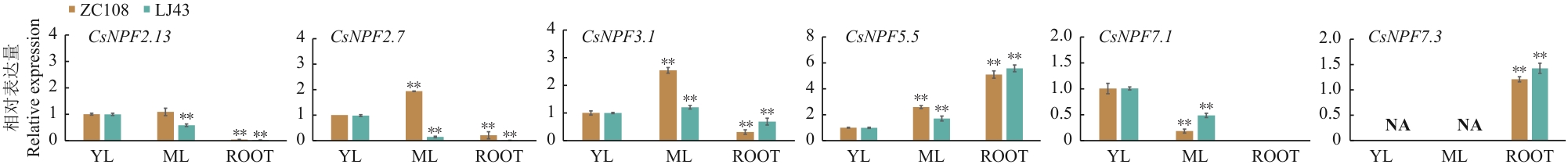
Fig. 5 Organizational characteristics analysis of CsNPFs genesDifferent tea varieties ‘Zhongcha 108’ (ZC108), ‘Longjing 43’ (LJ43), different tissue parts of the young leaf (YL), mature leaf (ML), root (ROOT). NA indicates that data is not detected. The error bars indicate the standard deviation among the three replicates. The data are three biological replicates ± SD. * P < 0.05, ** P < 0.01. The same below
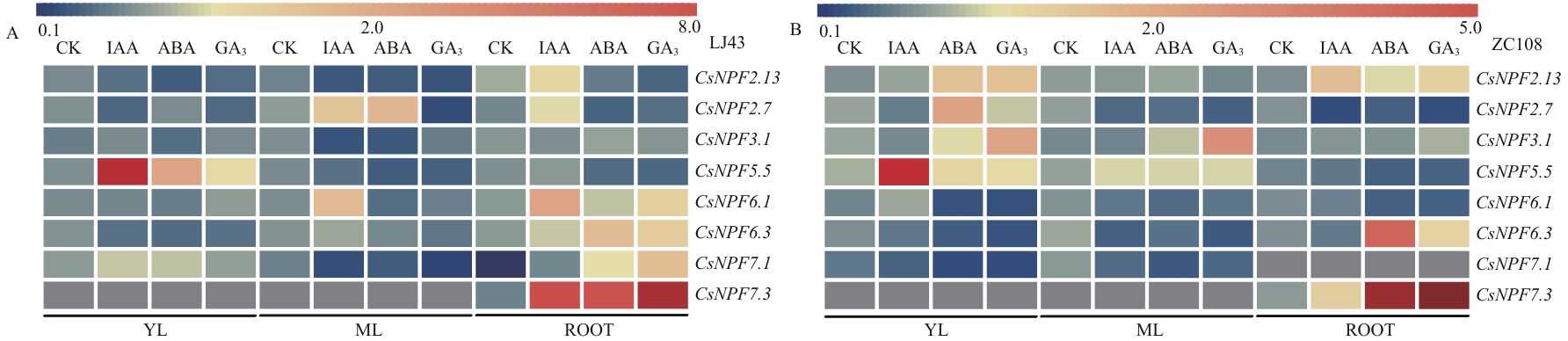
Fig. 6 Expressions of CsNPFs in different tissues under hormone treatmentThe average values of three biological replicates were used to generate a heat map using MeV software. The intensity value bars are shown above the heat map. Blue denotes low expression, and red denotes high expression. The grey color indicates that no data has been detected

Fig. 7 Expressions of CsNPFs genes in the leaves and roots under nitrate treatmentNitrogen starvation treatment for 10 d (-N), nitrogen addition 1 mol/L (low nitrogen, +LN), 4 mol/L (high nitrogen, +HN), respectively, for 2 h and 2 d
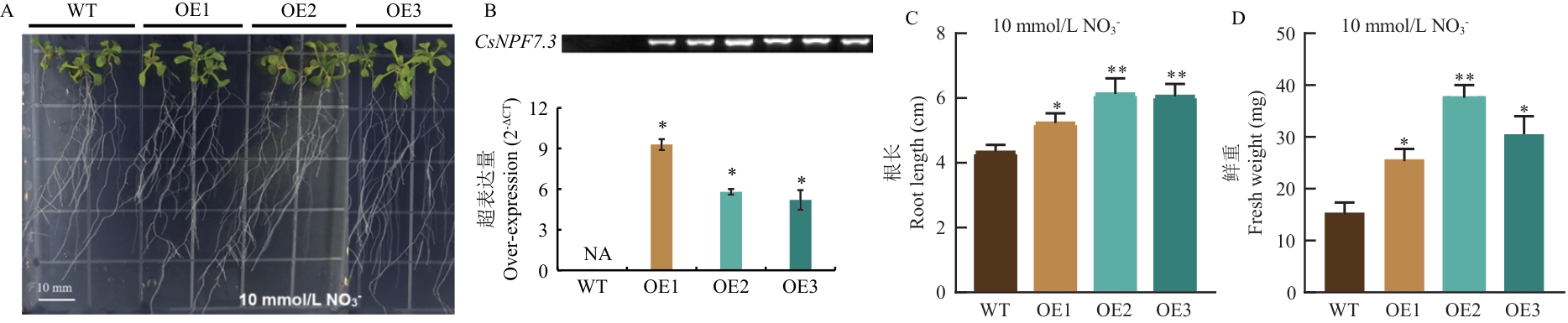
Fig. 8 Phenotypic difference analysis of CsNPF7.3 overexpressing lines (OE) compared with WT (Col-0) under different treatment conditionsA: Phenotype of Arabidopsis thaliana growth under 1/4 MS (10 mmol/L NO3-) conditions. B: Positive identification of overexpressing lines. C: Root lengths of different lines under 10 mmol/L NO3- conditions. D: Fresh weights of different lines under 10 mmol/L NO3- conditions. WT refers to the wild-type A. thaliana Col-0, and OE refers to the CsNPF7.3 transgenic overexpressing A. thaliana (OE1, OE2, and OE3 are 3 overexpressing lines). The same below
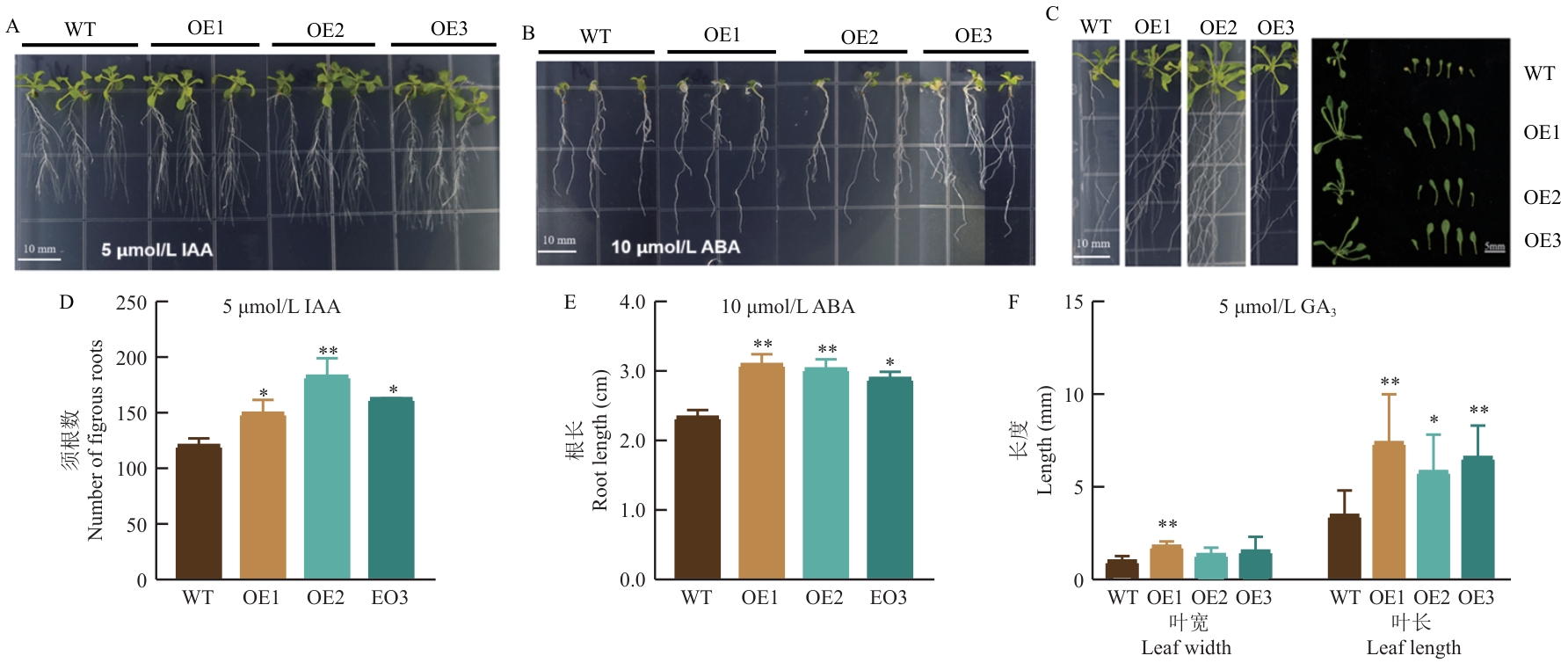
Fig. 9 Phenotypic difference analysis of CsNPF7.3 overexpressing lines (OE) compared with WT (Col-0) under different hormone treatment conditionsA: Phenotype of plant growth under 5 μmol/L IAA conditions. B: Phenotype of plant growth under 10 μmol/L ABA conditions. C: Phenotype of plant growth under 5 μmol/L GA3 conditions. D: The number of fibrous roots analysis among different lines under 5 μmol/L IAA conditions. E: Analysis of the difference in root length among plants under 10 μmol/L ABA conditions. F: Analysis of the difference in leaf width and leaf length among plants under 5 μmol/L GA3 conditions
| [1] | 袁青云, 贺巍, 苏会, 等. 植物硝酸根/多肽转运蛋白NPF家族功能研究进展 [J]. 植物生理学报, 2022, 58(10): 1840-1850. |
| Yuan QY, He W, Su H, et al. Research progress in nitrate/peptide transporter NPF family in plants [J]. Plant Physiol J, 2022, 58(10): 1840-1850. | |
| [2] | 黄慧梅, 高永康, 台玉莹, 等. 硝酸盐转运蛋白NRT2在植物中的功能及分子机制研究进展 [J]. 植物学报, 2023, 58(5): 783-798. |
| Huang HM, Gao YK, Tai YY, et al. Research advances in elucidating the function and molecular mechanism of the nitrate transporter 2 (NRT2) proteins in plants [J]. Chin Bull Bot, 2023, 58(5): 783-798. | |
| [3] | Alboresi A, Gestin C, Leydecker MT, et al. Nitrate, a signal relieving seed dormancy in Arabidopsis [J]. Plant Cell Environ, 2005, 28(4): 500-512. |
| [4] | 钱雨轩, 武占会, 刘宁, 等. 生菜NRT2/3的生物信息及功能的初步验证 [J]. 植物营养与肥料学报, 2024, 30(11): 2161-2180. |
| Qian YX, Wu ZH, Liu N, et al. Biological information of lettuce NRT2/3 and preliminary functional validation [J]. J Plant Nutr Fertil, 2024, 30(11): 2161-2180. | |
| [5] | 胡书婷, 王青. 植物氮代谢与氮素利用遗传基础研究进展 [J]. 山西农业科学, 2024, 52(6): 1-11. |
| Hu ST, Wang Q. Research progress on nitrogen metabolism and genetic basis of nitrogen utilization in plant [J]. J Shanxi Agric Sci, 2024, 52(6): 1-11. | |
| [6] | 尹卓然, 王驰, 轩栋栋, 等. 烟草硝酸盐转运蛋白NtNRT1.7基因的克隆、亚细胞定位及表达模式分析 [J]. 烟草科技, 2022, 55(5): 1-8. |
| Yin ZR, Wang C, Xuan DD, et al. Cloning, subcellular localization and expression pattern analysis of tobacco nitrate transporter NtNRT1.7 gene [J]. Tob Sci Technol, 2022, 55(5): 1-8. | |
| [7] | Fan SC, Lin CS, Hsu PK, et al. The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate [J]. Plant Cell, 2009, 21(9): 2750-2761. |
| [8] | Corratgé-Faillie C, Lacombe B. Substrate (un) specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins [J]. J Exp Bot, 2017, 68(12): 3107-3113. |
| [9] | 冯凯月, 赵鑫焱, 李子妍, 等. 植物响应盐碱胁迫的分子机制研究进展 [J]. 生物技术通报, 2024, 40(10): 122-138. |
| Feng KY, Zhao XY, Li ZY, et al. Research advances in the molecular mechanisms of plant response to saline-alkali stress [J]. Biotechnol Bull, 2024, 40(10): 122-138. | |
| [10] | Segonzac C, Boyer JC, Ipotesi E, et al. Nitrate efflux at the root plasma membrane: identification of an Arabidopsis excretion transporter [J]. Plant Cell, 2007, 19(11): 3760-3777. |
| [11] | Chiu CC, Lin CS, Hsia AP, et al. Mutation of a nitrate transporter, AtNRT1: 4, results in a reduced petiole nitrate content and altered leaf development [J]. Plant Cell Physiol, 2004, 45(9): 1139-1148. |
| [12] | Almagro A, Lin SH, Tsay YF. Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development [J]. Plant Cell, 2008, 20(12): 3289-3299. |
| [13] | Fang ZM, Bai GX, Huang WT, et al. The rice peptide transporter OsNPF7.3 is induced by organic nitrogen, and contributes to nitrogen allocation and grain yield [J]. Front Plant Sci, 2017, 8: 1338. |
| [14] | Wang J, Lu K, Nie HP, et al. Rice nitrate transporter OsNPF7.2 positively regulates tiller number and grain yield [J]. Rice, 2018, 11(1): 12. |
| [15] | Krapp A. Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces [J]. Curr Opin Plant Biol, 2015, 25: 115-122. |
| [16] | Krouk G, Crawford NM, Coruzzi GM, et al. Nitrate signaling: adaptation to fluctuating environments [J]. Curr Opin Plant Biol, 2010, 13(3): 265-272. |
| [17] | Tal I, Zhang Y, Jørgensen ME, et al. The Arabidopsis NPF3 protein is a GA transporter [J]. Nat Commun, 2016, 7: 11486. |
| [18] | Shimizu T, Kanno Y, Suzuki H, et al. Arabidopsis NPF4.6 and NPF5.1 control leaf stomatal aperture by regulating abscisic acid transport [J]. Genes, 2021, 12(6): 885. |
| [19] | Tang Z, Chen Y, Chen F, et al. OsPTR7 (OsNPF8.1), a putative peptide transporter in rice, is involved in dimethylarsenate accumulation in rice grain [J]. Plant Cell Physiol, 2017, 58(5): 904-913. |
| [20] | Niu HL, Wang JJ, Liao ZW, et al. Root-specific expression of CsNPF2.3 is involved in modulating fluoride accumulation in tea plant (Camellia sinensis) [J]. Hortic Res, 2025, 12(6): uhaf072. |
| [21] | Zhang F, Wang LY, Bai PX, et al. Identification of regulatory networks and hub genes controlling nitrogen uptake in tea plants [Camellia sinensis (L.) O. kuntze [J]. J Agric Food Chem, 2020, 68(8): 2445-2456. |
| [22] | 袁青云, 韩昱, 贺巍, 等. 茶树CsNPF6.1/6.3基因克隆及ABA转运功能验证 [J]. 园艺学报, 2024, 51(12): 2817-2828. |
| Yuan QY, Han Y, He W, et al. Cloning and ABA transport function analysis of CsNPF6.1/6.3 in tea plants [J]. Acta Hortic Sin, 2024, 51(12): 2817-2828. | |
| [23] | Muoki RC, Paul A, Kumari A, et al. An improved protocol for the isolation of RNA from roots of tea (Camellia sinensis (L.) O. Kuntze) [J]. Mol Biotechnol, 2012, 52(1): 82-88. |
| [24] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001, 25(4): 402-408. |
| [25] | Bent A. Arabidopsis thaliana floral dip transformation method [J]. Methods Mol Biol, 2006, 343: 87-103. |
| [26] | Zhang XR, Henriques R, Lin SS, et al. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method [J]. Nat Protoc, 2006, 1(2): 641-646. |
| [27] | 李婧, 左欣欣, 赵培伶, 等. 茶树高亲和硝酸盐转运蛋白家族基因NRT2的鉴定与表达 [J]. 应用与环境生物学报, 2022, 28(1): 50-56. |
| Li J, Zuo XX, Zhao PL, et al. Identification and expression analysis of high-affinity nitrate transporter family genes NRT2 in Camellia sinensis [J]. Chin J Appl Environ Biol, 2022, 28(1): 50-56. | |
| [28] | 马嘉俊, 吴英华, 李璿, 等. 白菜NPF基因家族成员的鉴定及其生物信息学分析 [J]. 河南农业科学, 2021, 50(9): 117-127. |
| Ma JJ, Wu YH, Li X, et al. Identification and bioinformatics analysis of NPF gene family members in Chinese Cabbage (Brassica rapa subsp. pekinensis) [J]. J Henan Agric Sci, 2021, 50(9): 117-127. | |
| [29] | Wen J, Li PF, Ran F, et al. Genome-wide characterization, expression analyses, and functional prediction of the NPF family in Brassica napus [J]. BMC Genomics, 2020, 21(1): 871. |
| [30] | Tsay YF, Chiu CC, Tsai CB, et al. Nitrate transporters and peptide transporters [J]. FEBS Lett, 2007, 581(12): 2290-2300. |
| [31] | Parker JL, Newstead S. Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1 [J]. Nature, 2014, 507(7490): 68-72. |
| [32] | 汪芳珍. 荒漠植物沙芥NAXT家族基因的克隆及其成员PcNPF2.7的功能分析 [D]. 兰州: 兰州大学, 2021. |
| Wang FZ. Cloning of NAXT family genes from desert plant Pugionium cornutum and functional analysis of its member PcNPF2.7 [D]. Lanzhou: Lanzhou University, 2021. | |
| [33] | Léran S, Garg B, Boursiac Y, et al. AtNPF5.5, a nitrate transporter affecting nitrogen accumulation in Arabidopsis embryo [J]. Sci Rep, 2015, 5: 7962. |
| [34] | Cui YN, Li XT, Yuan JZ, et al. Nitrate transporter NPF7.3/NRT1.5 plays an essential role in regulating phosphate deficiency responses in Arabidopsis [J]. Biochem Biophys Res Commun, 2019, 508(1): 314-319. |
| [35] | Lin SH, Kuo HF, Canivenc G, et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport [J]. Plant Cell, 2008, 20(9): 2514-2528. |
| [36] | 郑冬超, 夏新莉, 尹伟伦. 生长素促进拟南芥AtNRT1.1基因表达增强硝酸盐吸收 [J]. 北京林业大学学报, 2013, 35(2): 80-85. |
| Zheng DC, Xia XL, Yin WL. Auxin promotes nitrate uptake by up-regulating AtNRT1.1gene transcript level in Arobidopsis thaliana [J]. J Beijing For Univ, 2013, 35(2): 80-85. | |
| [37] | Chen HF, Zhang Q, Wang XR, et al. Nitrogen form-mediated ethylene signal regulates root-to-shoot K+ translocation via NRT1.5 [J]. Plant Cell Environ, 2021, 44(12): 3806-3818. |
| [38] | Guo FQ, Wang RC, Crawford NM. The Arabidopsis dual‐affinity nitrate transporter gene AtNRT1.1 (CHL1) is regulated by auxin in both shoots and roots [J]. J Exp Bot, 2002, 53(370): 835-844. |
| [1] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [2] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [3] | GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses [J]. Biotechnology Bulletin, 2025, 41(9): 82-93. |
| [4] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| [5] | ZHANG Yong-yan, GUO Si-jian, LI Jing, HAO Si-yi, LI Rui-de, LIU Jia-peng, CHENG Chun-zhen. Gene Cloning and Functional Analysis of the Anthocyanin-related VcGSTF19 Gene in Blueberry (Vaccinium corymbosum L.) [J]. Biotechnology Bulletin, 2025, 41(9): 139-146. |
| [6] | BAI Yu-guo, LI Wan-di, LIANG Jian-ping, SHI Zhi-yong, LU Geng-long, LIU Hong-jun, NIU Jing-ping. Growth-promoting Mechanism of Trichoderma harzianum T9131 on Astragalus membranaceus Seedlings [J]. Biotechnology Bulletin, 2025, 41(8): 175-185. |
| [7] | HUA Wen-ping, LIU Fei, HAO Jia-xin, CHEN Chen. Identification and Expression Patterns Analysis of ADH Gene Family in Salvia miltiorrhiza [J]. Biotechnology Bulletin, 2025, 41(8): 211-219. |
| [8] | LI Kai-jie, WU Yao, LI Dan-dan. Cloning of Gene CtbHLH128 in Safflower and Response Function Regulating Drought Stress [J]. Biotechnology Bulletin, 2025, 41(8): 234-241. |
| [9] | REN Rui-bin, SI Er-jing, WAN Guang-you, WANG Jun-cheng, YAO Li-rong, ZHANG Hong, MA Xiao-le, LI Bao-chun, WANG Hua-jun, MENG Ya-xiong. Identification and Expression Analysis of GH17 Gene Family of Pyrenophora graminea [J]. Biotechnology Bulletin, 2025, 41(8): 146-154. |
| [10] | KANG Qin, WANG Xia, SHEN Ming-yang, XU Jing-tian, CHEN Shi-lan, LIAO Ping-yang, XU Wen-zhi, WU Wei, XU Dong-bei. Cloning and Expression Analysis of the UV-B Receptor Gene McUVR8 in Mentha canadensis L. [J]. Biotechnology Bulletin, 2025, 41(8): 255-266. |
| [11] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| [12] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [13] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [14] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [15] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||