








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (7): 214-225.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0048
Previous Articles Next Articles
GONG Yu-han( ), CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian(
), CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian( )
)
Received:2025-01-12
Online:2025-07-26
Published:2025-07-22
Contact:
LIU Shuo-qian
E-mail:3166693974@qq.com;shuoqianliu@hunau.edu.cn
GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant[J]. Biotechnology Bulletin, 2025, 41(7): 214-225.
| 基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| CsTRB1 | TTTGAAGGAGCCTCGTGGG | GTTGCTGCCAACAGCCTTTC |
| CsTRB2 | TGTTGATCTCAAGGACAAATGGAG | ATCCTGTTTAGGGGCAGGCT |
| CsTRB3 | GGCACCTGCCAACTTCAAAC | CCCTCTGCCTTCCCTCTAGT |
| CsTRB4 | TGGTTGTGGCAAAAACACCG | TGCCTCCTTAACGGCTTCAG |
| CsTRB5 | AAAAGCAGAAGTGGACGGCT | CCATGTTCCGCCATTTGTCC |
| CsTRB6 | CTGAATTTGCTCTTTCTCTCACCC | ATTTTGACCATTGGTGAGTGGG |
| CsTRB7 | ATCCGGAATTTAGTGGCGTCT | CTCCATTTGTCCTTGAGATCAACA |
| CsGAPDH | TTGGCATCGTTGAGGGTCT | CAGTGGGAACACGGAAAGC |
| PCR-CsTRB1 | ATGGTGGTCGCCTCCGGA | CTAAATCCAAATGCTACTGAAGGCT |
Table 1 Primer information
| 基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| CsTRB1 | TTTGAAGGAGCCTCGTGGG | GTTGCTGCCAACAGCCTTTC |
| CsTRB2 | TGTTGATCTCAAGGACAAATGGAG | ATCCTGTTTAGGGGCAGGCT |
| CsTRB3 | GGCACCTGCCAACTTCAAAC | CCCTCTGCCTTCCCTCTAGT |
| CsTRB4 | TGGTTGTGGCAAAAACACCG | TGCCTCCTTAACGGCTTCAG |
| CsTRB5 | AAAAGCAGAAGTGGACGGCT | CCATGTTCCGCCATTTGTCC |
| CsTRB6 | CTGAATTTGCTCTTTCTCTCACCC | ATTTTGACCATTGGTGAGTGGG |
| CsTRB7 | ATCCGGAATTTAGTGGCGTCT | CTCCATTTGTCCTTGAGATCAACA |
| CsGAPDH | TTGGCATCGTTGAGGGTCT | CAGTGGGAACACGGAAAGC |
| PCR-CsTRB1 | ATGGTGGTCGCCTCCGGA | CTAAATCCAAATGCTACTGAAGGCT |
基因 ID Gene ID | 基因名 Gene name | 氨基酸数Number of amino acids | 蛋白质分子量 Molecular weight (Da) | 等电点Theoretical pI | 不稳定系数 Instability index | 脂肪指数Aliphatic index | 亲水性 Grand of hydropathicity | 亚细胞定位 Subcellular localization prediction |
|---|---|---|---|---|---|---|---|---|
| CSS0029061 | CsTRB1 | 294 | 32 011.53 | 8.56 | 34.83 | 82.52 | -0.497 | 细胞核 |
| CSS0034494 | CsTRB2 | 302 | 33 233.12 | 9.52 | 46.42 | 82.28 | -0.530 | 细胞核 |
| CSS0012155 | CsTRB3 | 300 | 32 813.69 | 9.56 | 49.24 | 81.57 | -0.470 | 细胞核 |
| CSS0030254 | CsTRB4 | 280 | 30 857.84 | 9.16 | 40.08 | 69.79 | -0.688 | 细胞核 |
| CSS0035646 | CsTRB5 | 264 | 29 439.74 | 9.71 | 37.99 | 81.40 | -0.616 | 细胞核 |
| CSS0046266 | CsTRB6 | 264 | 29 439.74 | 9.71 | 37.99 | 81.40 | -0.616 | 细胞核 |
| CSS0022401 | CsTRB7 | 302 | 33 234.06 | 9.52 | 46.42 | 80.99 | -0.557 | 细胞核 |
Table 2 Physicochemical properties of CsTRBs proteins in Camellia sinensis
基因 ID Gene ID | 基因名 Gene name | 氨基酸数Number of amino acids | 蛋白质分子量 Molecular weight (Da) | 等电点Theoretical pI | 不稳定系数 Instability index | 脂肪指数Aliphatic index | 亲水性 Grand of hydropathicity | 亚细胞定位 Subcellular localization prediction |
|---|---|---|---|---|---|---|---|---|
| CSS0029061 | CsTRB1 | 294 | 32 011.53 | 8.56 | 34.83 | 82.52 | -0.497 | 细胞核 |
| CSS0034494 | CsTRB2 | 302 | 33 233.12 | 9.52 | 46.42 | 82.28 | -0.530 | 细胞核 |
| CSS0012155 | CsTRB3 | 300 | 32 813.69 | 9.56 | 49.24 | 81.57 | -0.470 | 细胞核 |
| CSS0030254 | CsTRB4 | 280 | 30 857.84 | 9.16 | 40.08 | 69.79 | -0.688 | 细胞核 |
| CSS0035646 | CsTRB5 | 264 | 29 439.74 | 9.71 | 37.99 | 81.40 | -0.616 | 细胞核 |
| CSS0046266 | CsTRB6 | 264 | 29 439.74 | 9.71 | 37.99 | 81.40 | -0.616 | 细胞核 |
| CSS0022401 | CsTRB7 | 302 | 33 234.06 | 9.52 | 46.42 | 80.99 | -0.557 | 细胞核 |

Fig. 1 Amino acid sequence alignment of CsTRB from tea plant with other TRB family proteinsThe MYB domain is marked with a red line, the histone H1/5 domain is marked with a yellow line, and the coil domain is marked with a blue line. Cs: Camellia sinensis. At: Arabidopsis thaliana. Os: Oryza sativa. Zm: Zea mays
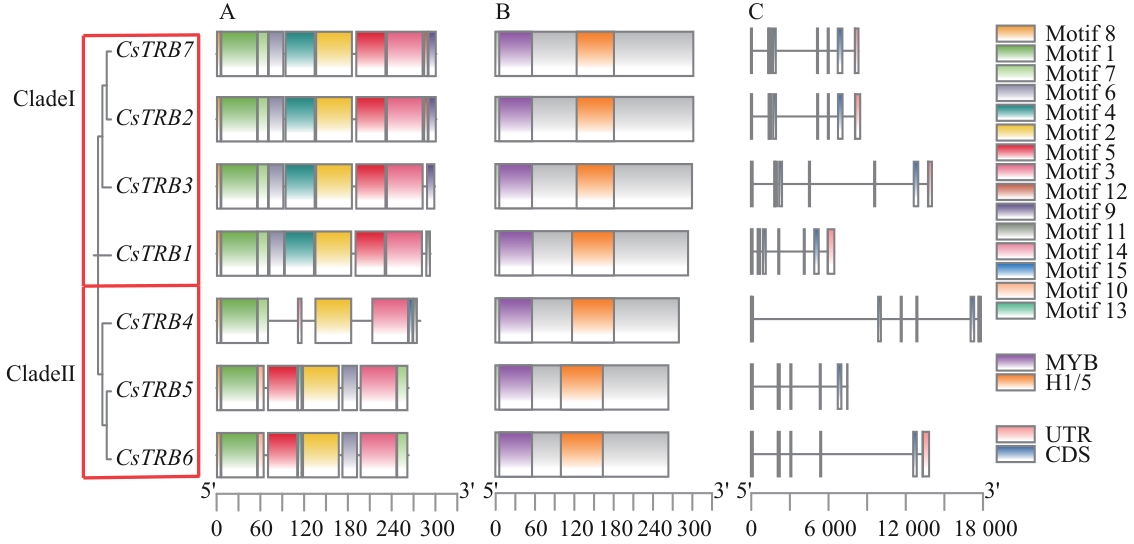
Fig. 4 Motif, CDD and structural analysis of TRB family genes in C. sinensisA: Identification based on phylogenetic relationships and domain, CsTRBs were classified into two subgroups. B: Composition of conserved motifs within CsTRB proteins. Boxes of different colors indicate distinct domains. C: Gene structures of CsTRBs. Blue rectangles denote CDS or exons, while gray lines indicate introns
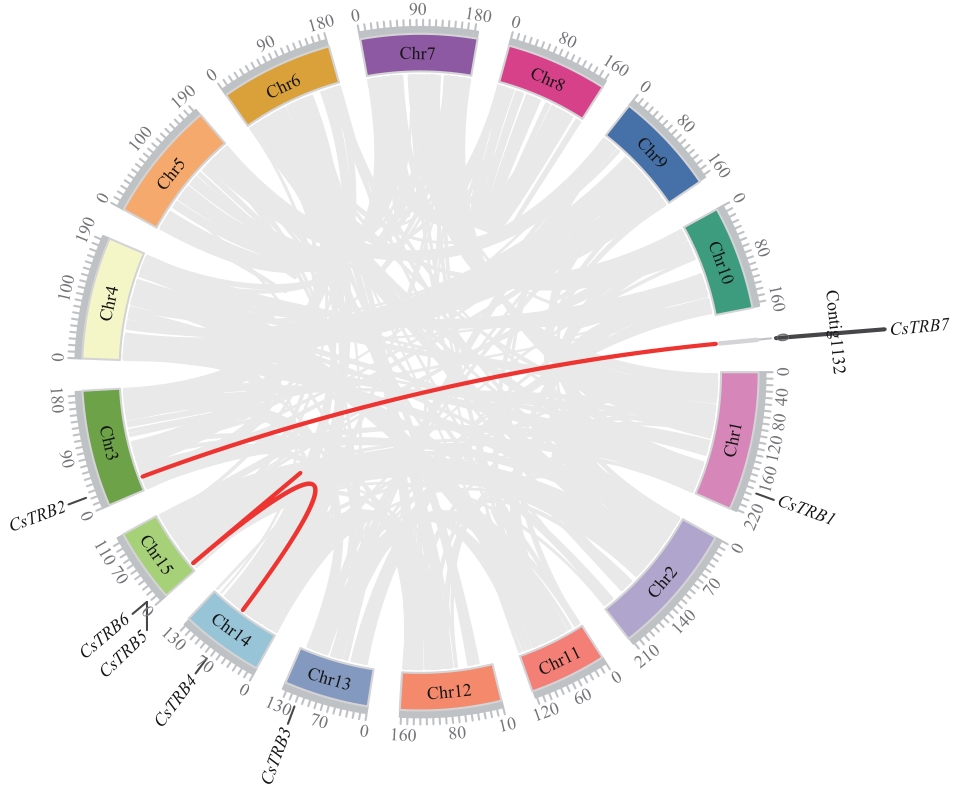
Fig. 5 Chromosomal localization and inter-chromosomal associations of CsTRB genesIn the background, gray lines indicate all the synsynblocks in the tea plant genome, while red lines indicate the synsynbnlor gene pairs

Fig. 6 Synteny analysis of TRB gene family between C. sinensis and A. thalianThe gray lines in the background depict syntenic blocks between the tea plant and Arabidopsis, while blue lines indicate syntenic TRB gene pairs

Fig. 9 Transcriptome expression profile of CsTRB genesA, C, D, and E: Indicate the expression analysis of CsTRB gene in different tissues, 200 mmol/L NaCl, 25% PEG and 100 mmol/L MeJA stress, the data were converted by log2 (FPKM+1), the red square indicates that the gene was up-regulated, and the blue square indicates that the gene was down-regulated. B: Expression analysis of CsTRB gene in tea plant under low temperature acclimation (CK: Non-acclimated at 25 ℃; Cold1-6 h: fully acclimated at 10 ℃ for 6 h; Cold1-7 d: 10 ℃/4 ℃ at day/night for 7 d; Cold2-7 d: 4 ℃/0 ℃ at day/night for 7 d; ColdDA-7 d: recovering under 25 ℃ for 7 d)

Fig. 10 Expression profile of CsTRB gene under different abiotic stressesA: Expression profile of CsTRBs gene under low temperature stress. B: Expression profile of CsTRBs gene under 100 μmol/L ABA treatment. C: Expression profile of the CsTRBs gene under 100 μmol/L IAA treatment. The error bars indicate the standard deviation of 3 independent biological replicates. Different lowercase letters indicate significant differences at different time points of the same treatment (P<0.05)
| [1] | Riha K, Shippen DE. Telomere structure, function and maintenance in Arabidopsis [J]. Chromosome Res, 2003, 11(3): 263-275. |
| [2] | Bilaud T, Koering CE, Binet-Brasselet E, et al. The telobox, a MYB-related telomeric DNA binding motif found in proteins from yeast, plants and human [J]. Nucleic Acids Res, 1996, 24(7): 1294-1303. |
| [3] | Peška V, Schrumpfová PP, Fajkus J. Using the telobox to search for plant telomere binding proteins [J]. Curr Protein Pept Sci, 2011, 12(2): 75-83. |
| [4] | Kuchar M, Fajkus J. Interactions of putative telomere-binding proteins in Arabidopsis thaliana: identification of functional TRF2 homolog in plants [J]. FEBS Lett, 2004, 578(3): 311-315. |
| [5] | Schrumpfová P, Kuchar M, Miková G, et al. Characterization of two Arabidopsis thaliana myb-like proteins showing affinity to telomeric DNA sequence [J]. Genome, 2004, 47(2): 316-324. |
| [6] | Marian CO, Bordoli SJ, Goltz M, et al. The maize Single myb histone 1 gene, Smh1, belongs to a novel gene family and encodes a protein that binds telomere DNA repeats in vitro [J]. Plant Physiol, 2003, 133(3): 1336-1350. |
| [7] | Amiard S, Feit L, Simon L, et al. Distinct clades of telomere repeat binding transcriptional regulators interplay to regulate plant development [J/OL]. bioRxiv, 2023. DOI:10.1101/2023.08.16.553498 . |
| [8] | He Q, Chen L, Xu Y, et al. Identification of centromeric and telomeric DNA-binding proteins in rice [J]. Proteomics, 2013, 13(5): 826-832. |
| [9] | Yang SW, Kim DH, Lee JJ, et al. Expression of the telomeric repeat binding factor gene NgTRF1 is closely coordinated with the cell division program in tobacco BY-2 suspension culture cells [J]. J Biol Chem, 2003, 278(24): 21395-21407. |
| [10] | 张冬杰, 燕丽萍, 吴其超, 等. 绒毛白蜡smh1基因编码区的克隆及序列分析 [J]. 中国农学通报, 2017, 33(15): 49-55. |
| Zhang DJ, Yan LP, Wu QC, et al. Cloning and sequence analysis of Fraxinus velutina smh1 gene coding region [J]. Chin Agric Sci Bull, 2017, 33(15): 49-55. | |
| [11] | Mozgová I, Schrumpfová PP, Hofr C, et al. Functional characterization of domains in AtTRB1, a putative telomere-binding protein in Arabidopsis thaliana [J]. Phytochemistry, 2008, 69(9): 1814-1819. |
| [12] | Hofr C, Sultesová P, Zimmermann M, et al. Single-myb-histone proteins from Arabidopsis thaliana: a quantitative study of telomere-binding specificity and kinetics [J]. Biochem J, 2009, 419(1): 221-230. |
| [13] | Schrumpfová PP, Vychodilová I, Dvořáčková M, et al. Telomere repeat binding proteins are functional components of Arabidopsis telomeres and interact with telomerase [J]. Plant J, 2014, 77(5): 770-781. |
| [14] | Chen YH, Yang XY, He K, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family [J]. Plant Mol Biol, 2006, 60(1): 107-124. |
| [15] | An JP, Xu RR, Liu X, et al. Jasmonate induces biosynthesis of anthocyanin and proanthocyanidin in apple by mediating the JAZ1-TRB1-MYB9 complex [J]. Plant J, 2021, 106(5): 1414-1430. |
| [16] | Zhou Y, Wang YJ, Krause K, et al. Telobox motifs recruit CLF/SWN-PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis [J]. Nat Genet, 2018, 50(5): 638-644. |
| [17] | Hong JP, Byun MY, Koo DH, et al. Suppression of RICE TELOMERE BINDING PROTEIN 1 results in severe and gradual developmental defects accompanied by genome instability in rice [J]. Plant Cell, 2007, 19(6): 1770-1781. |
| [18] | Procházková Schrumpfová P, Schořová Š, Fajkus J. Telomere- and telomerase-associated proteins and their functions in the plant cell [J]. Front Plant Sci, 2016, 7: 851. |
| [19] | 莫晓丽, 黄亚辉. 茶树主要逆境胁迫反应及其适应逆境的生理机制 [J]. 茶叶学报, 2021, 62(4): 185-190. |
| Mo XL, Huang YH. Responses and resistance mechanisms of tea plants to stresses—a review [J]. Acta Tea Sin, 2021, 62(4): 185-190. | |
| [20] | 王鹏杰, 杨江帆, 张兴坦, 等. 茶树基因组与测序技术的研究进展 [J]. 茶叶科学, 2021, 41(6): 743-752. |
| Wang PJ, Yang JF, Zhang XT, et al. Research advance of tea plant genome and sequencing technologies [J]. J Tea Sci, 2021, 41(6): 743-752. | |
| [21] | El-Gebali S, Mistry J, Bateman A, et al. The pfam protein families database in 2019 [J]. Nucleic Acids Res, 2019, 47(D1): D427-D432. |
| [22] | Potter SC, Luciani A, Eddy SR, et al. HMMER web server: 2018 update [J]. Nucleic Acids Res, 2018, 46(w1): W200-W204. |
| [23] | Larkin MA, Blackshields G, Brown NP, et al. Clustal W and clustal X version 2.0 [J]. Bioinformatics, 2007, 23(21): 2947-2948. |
| [24] | Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11 [J]. Mol Biol Evol, 2021, 38(7): 3022-3027. |
| [25] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [26] | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching [J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| [27] | Wang YP, Tang HB, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity [J]. Nucleic Acids Res, 2012, 40(7): e49. |
| [28] | Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics [J]. Genome Res, 2009, 19(9): 1639-1645. |
| [29] | Wang LQ, Guo K, Li Y, et al. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice [J]. BMC Plant Biol, 2010, 10: 282. |
| [30] | Pontius J, Richelle J, Wodak SJ. Deviations from standard atomic volumes as a quality measure for protein crystal structures [J]. J Mol Biol, 1996, 264(1): 121-136. |
| [31] | Delano WL. Pymol: An open-source molecular graphics tool [J]. CCP4 Newsletter on Protein Crystallography, 2002, 40(1): 82-92. |
| [32] | Swift ML. GraphPad prism, data analysis, and scientific graphing [J]. J Chem Inf Comput Sci, 1997, 37(2): 411-412. |
| [33] | Wang L, Ding XL, Gao YQ, et al. Genome-wide identification and characterization of GRAS genes in soybean (Glycine max) [J]. BMC Plant Biol, 2020, 20(1): 415. |
| [34] | Xu GX, Guo CC, Shan HY, et al. Divergence of duplicate genes in exon-intron structure [J]. Proc Natl Acad Sci USA, 2012, 109(4): 1187-1192. |
| [35] | Roy SW, Fedorov A, Gilbert W. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain [J]. Proc Natl Acad Sci USA, 2003, 100(12): 7158-7162. |
| [36] | Rose AB. Introns as gene regulators: a brick on the accelerator [J]. Front Genet, 2019, 9: 672. |
| [37] | Jeffares DC, Penkett CJ, Bähler J. Rapidly regulated genes are intron poor [J]. Trends Genet, 2008, 24(8): 375-378. |
| [38] | Chen HY, Hsieh EJ, Cheng MC, et al. ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element [J]. New Phytol, 2016, 211(2): 599-613. |
| [39] | Zhou Y, Hartwig B, James GV, et al. Complementary activities of telomere repeat binding proteins and polycomb group complexes in transcriptional regulation of target genes [J]. Plant Cell, 2016, 28(1): 87-101. |
| [40] | Kusová A, Steinbachová L, Přerovská T, et al. Completing the TRB family: newly characterized members show ancient evolutionary origins and distinct localization, yet similar interactions [J]. Plant Mol Biol, 2023, 112(1/2): 61-83. |
| [1] | ZHANG Xue-qiong, PAN Su-jun, LI Wei, DAI Liang-ying. Research Progress of Plant Phosphate Transporters in the Response to Stress [J]. Biotechnology Bulletin, 2025, 41(7): 28-36. |
| [2] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [3] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [4] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [5] | LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 181-192. |
| [6] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [7] | WANG Miao-miao, ZHAO Xiang-long, WANG Zhao-ming, LIU Zhi-peng, YAN Long-feng. Identification of TCP Gene Family in Medicago ruthenica and Their Expression Pattern Analysis under Drought Stress [J]. Biotechnology Bulletin, 2025, 41(6): 179-190. |
| [8] | WU Hao, DONG Wei-feng, HE Zi-tian, LI Yan-xiao, XIE Hui, SUN Ming-zhe, SHEN Yang, SUN Xiao-li. Genome-wide Identification and Expression Analysis of the Rice BXL Gene Family [J]. Biotechnology Bulletin, 2025, 41(6): 87-98. |
| [9] | QU Mei-ling, ZHOU Si-min, ZHANG Jing-yu, HE Jia-wei, ZHU Jia-yuan, LIU Xiao-rong, TONG Qiao-zhen, ZHOU Ri-bao, LIU Xiang-dan. Identification and Expression Analysis of bHLH Transcription Gene Family in Lonicera macranthoides [J]. Biotechnology Bulletin, 2025, 41(6): 256-268. |
| [10] | HUANG Dan, PENG Bing-yang, ZHANG Pan-pan, JIAO Yue, LYU Jia-bin. Identification of HD-Zip Gene Family in Camellia oleifera and Analysis of Its Expression under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(6): 191-207. |
| [11] | LIU Xin, WANG Jia-wen, LI Jin-wei, MOU Ce, YANG Pan-pan, MING Jun, XU Lei-feng. Cloning and Expression Analysis of Three LdBBXs in Lilium davidii var. willmottiae [J]. Biotechnology Bulletin, 2025, 41(5): 186-196. |
| [12] | PENG Shao-zhi, WANG Deng-ke, ZHANG Xiang, DAI Xiong-ze, XU Hao, ZOU Xue-xiao. Cloning, Expression Characteristics and Functional Verification of the Pepper CaFD1 Gene [J]. Biotechnology Bulletin, 2025, 41(5): 153-164. |
| [13] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| [14] | LUO Si-fang, ZHANG Zu-ming, XIE Li-fang, GUO Zi-jing, CHEN Zhao-xing, YANG Yue-hua, YAN Xiang, ZHANG Hong-ming. Genome-wide Identification of GATA Gene Family of Jindou Kumquat (Fortunella hindsii) and Their Expression Analysis in Fruit Development [J]. Biotechnology Bulletin, 2025, 41(5): 218-230. |
| [15] | LI Zhi-qiang, LUO Zheng-qian, XU Lin-li, ZHOU Guo-hui, QU Si-yu, LIU En-liang, XU Dong-ting. Identification of the Soybean R2R3-MYB Gene Family Based on the T2T Genome and Their Expression Analysis under Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 141-152. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||