








生物技术通报 ›› 2022, Vol. 38 ›› Issue (3): 59-68.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1315
收稿日期:2021-10-20
出版日期:2022-03-26
发布日期:2022-04-06
作者简介:王晓丽,女,硕士研究生,研究方向:大豆遗传育种;E-mail: 基金资助:
WANG Xiao-li( ), QIN Jie, WANG Min, WANG Li-xiang, DU Wei-jun(
), QIN Jie, WANG Min, WANG Li-xiang, DU Wei-jun( )
)
Received:2021-10-20
Published:2022-03-26
Online:2022-04-06
摘要:
山西作为大豆起源地之一,根瘤菌资源丰富。为了进一步挖掘山西省的大豆根瘤菌资源,筛选出与当地主栽大豆品种共生固氮效果好的根瘤菌。本实验在大同广灵、太原清徐、晋中太谷三地采集不同大豆品种的根瘤,经过多次划线,对分离纯化出的单菌落进行菌落形态、刚果红染色、BTB染色表型鉴定;16S rDNA、nodA和nifH基因型鉴定。利用BOX-PCR对所有根瘤菌进行电泳检测,去除条带完全一致的菌株,将目的条带不一致的菌在Bionumerics@7.5软件中聚类分析。选代表性菌株接种到晋大88号和科丰1号两个大豆材料,筛选匹配性好的菌株。共分离出203株菌,其中187株根瘤菌可以扩增出nodA、nifH基因,172株快生型根瘤菌,属于S. fredii;15株慢生型根瘤菌,分别属于B. diazoefficiens和 B. daqingense。BOX-PCR将85株条带不一致的根瘤菌分为四大类群,以70%为界限共分为16类。筛选出与晋大88号匹配性较好的根瘤菌GL11和GL43,其地上部分干重较接种USDA110分别增加38.5%、30.1%;根干重分别增加22.2%、27.8%;根瘤鲜重分别增加24.3%、41.5%;根瘤干重分别增加36.6%、31.0%。筛选出与科丰1号匹配性较好的根瘤菌TG37,其株高、地上干重、根干重、根瘤鲜重、根瘤干重和根瘤数较接种USDA110分别增加8.1%、15.0%、28.6%、27.3%、44.3%和60.6%。山西省根瘤菌资源丰富,种类多样,S. fredii为优势种,此外还分离出两类慢生型根瘤菌B. diazoefficiens和 B. daqingense,丰富了根瘤菌资源。筛选出共生匹配性好的根瘤菌为大豆生产实践奠定了物质基础和理论依据。
王晓丽, 秦杰, 王敏, 王利祥, 杜维俊. 山西大豆根瘤菌的分离、鉴定及共生匹配性筛选[J]. 生物技术通报, 2022, 38(3): 59-68.
WANG Xiao-li, QIN Jie, WANG Min, WANG Li-xiang, DU Wei-jun. Isolation,Identification and Symbiotic Matching of Soybean Rhizobia from Shanxi Province[J]. Biotechnology Bulletin, 2022, 38(3): 59-68.
| Primer name | Primer sequence(5'-3') | Literature source |
|---|---|---|
| nifH-F | AAAGGYGGWATCGGYAARTCCACCAC | [12] |
| nifH-R | TTGTTSGCSGCRTACATSGCCATCAT | |
| nodA-F | TGCRGTGGAATNTRNNCTGGGAAA | [13] |
| nodA-R | GGNCCGTCRTCRAAWGTCATGTA | |
| 16S rDNA-F | AGAGTTTGATCCTGGCTCAG | [14] |
| 16S rDNA-R | GGTTACCTTGTTACGACTT |
表1 根瘤菌基因鉴定所用引物
Table 1 Primers for gene identification of rhizobia
| Primer name | Primer sequence(5'-3') | Literature source |
|---|---|---|
| nifH-F | AAAGGYGGWATCGGYAARTCCACCAC | [12] |
| nifH-R | TTGTTSGCSGCRTACATSGCCATCAT | |
| nodA-F | TGCRGTGGAATNTRNNCTGGGAAA | [13] |
| nodA-R | GGNCCGTCRTCRAAWGTCATGTA | |
| 16S rDNA-F | AGAGTTTGATCCTGGCTCAG | [14] |
| 16S rDNA-R | GGTTACCTTGTTACGACTT |

图1 根瘤菌表型鉴定 A:根瘤菌的单菌落形态图;B:快生型根瘤菌BTB染色图;C:快生型根瘤菌BTB染色图
Fig. 1 Phenotype identification of rhizobia A:A single colony morphology of rhizobia. B:A BTB staining image of fast-growing rhizobia. C:A BTB staining image of fast-growing rhizobia
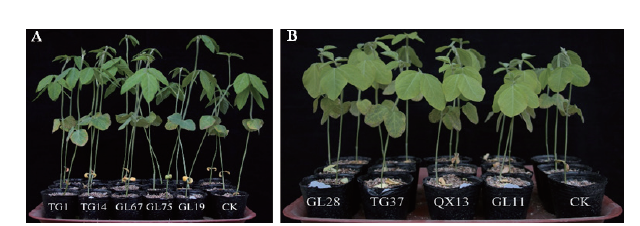
图9 根瘤菌与大豆的匹配性实验的大豆植株长势 A:晋大88号接菌后的植株长势;B:科丰1号接菌后的植株长势
Fig. 9 Soybean plant growth in the matching experiment between rhizobia and soybean A:The plant growth after inoculation of Jinda 88. B:The plant growth after inoculation of Kefeng 1

图10 根瘤菌与大豆的匹配性实验的根瘤图片 GL17-1、GL17-2和GL17-3分别是接种晋大88大豆根瘤菌GL17的3个重复;GL11-1、GL11-2和GL11-3分别是接种科丰1号大豆根瘤菌GL11的3个重复。
Fig. 10 Picture of nodules in the matching experiment between rhizobia and soybean GL17-1,GL17-2 and GL17-3 are three replicates of inoculating soybean Jinda 88 with Rhizobium GL17,respectively;GL11-1,GL11-2 and GL11-3 are three replicates of inoculating soybean Kefeng 1 with Rhizobium GL11,respectively
| Rhizobium | Jinda 88 | Kefeng 1 | |||||
|---|---|---|---|---|---|---|---|
| Plant height/cm | Shoot dry weight/g | Root dry weight/g | Plant height/cm | Shoot dry weight/g | Root dry weight/g | ||
| CK | 26.65±4.78 c | 0.23±0.07 c | 0.19±0.06 abc | 19.55±1.93 abcd | 0.18±0.02 bcde | 0.15±0.01 abc | |
| USDA110 | 31.40±2.91 abc | 0.26±0.04 bc | 0.18±0.02 abc | 19.83±0.45 abcd | 0.20±0.02 abcd | 0.14±0.00 bc | |
| TG1 | 30.15±4.16 abc | 0.28±0.05 abc | 0.19±0.01 abc | 21.80±2.17 a | 0.20±0.03 abc | 0.16±0.02 ab | |
| TG14 | 33.02±3.04 abc | 0.26±0.05 bc | 0.20±0.02 abc | 19.30±1.06 abcd | 0.16±0.03 cde | 0.12±0.02 c | |
| TG20 | 32.10±5.05 abc | 0.25±0.04 c | 0.19±0.02 abc | 16.40±1.67 d | 0.15±0.02 de | 0.12±0.01 c | |
| TG37 | 32.73±3.83 abc | 0.31±0.01 abc | 0.23±0.03 a | 21.43±2.35 ab | 0.23±0.01 a | 0.18±0.02 a | |
| TG53 | 34.45±4.74 abc | 0.28±0.04 abc | 0.21±0.03 abc | 20.37±1.29 abc | 0.17±0.02 bcde | 0.13±0.02 bc | |
| TG79 | 28.68±1.55 bc | 0.26±0.06 bc | 0.19±0.04 abc | 20.13±1.96 abc | 0.18±0.04 bcde | 0.14±0.03 bc | |
| GL11 | 30.63±5.23 abc | 0.36±0.02 a | 0.22±0.02 ab | 17.20±1.57 cd | 0.19±0.01 abcde | 0.14±0.00 bc | |
| GL16 | 31.77±2.96 abc | 0.25±0.03 c | 0.16±0.00 bc | 19.37±0.85 abcd | 0.18±0.01 bcde | 0.12±0.01 c | |
| GL17 | 32.33±5.72 abc | 0.31±0.07 abc | 0.19±0.04 abc | 18.23±2.41 bcd | 0.17±0.01 bcde | 0.12±0.02 c | |
| GL19 | 33.03±1.76 abc | 0.28±0.03 abc | 0.22±0.01 ab | 20.93±0.71 ab | 0.18±0.02 bcde | 0.13±0.01 bc | |
| GL28 | 34.60±2.25 ab | 0.30±0.02 abc | 0.21±0.03 ab | 19.67±1.11 abcd | 0.20±0.02 abc | 0.14±0.02 bc | |
| GL40 | 28.87±8.18 bc | 0.31±0.05 abc | 0.15±0.07 c | 16.97±2.30 cd | 0.16±0.03 cde | 0.13±0.01 c | |
| GL43 | 33.67±4.53 abc | 0.34±0.09 ab | 0.23±0.06 a | 19.47±2.39 abcd | 0.21±0.01 ab | 0.15±0.01 abc | |
| GL67 | 35.53±4.24 ab | 0.29±0.04 abc | 0.21±0.01 ab | 17.93±2.57 bcd | 0.16±0.00 cde | 0.14±0.02 bc | |
| GL75 | 36.97±0.47 a | 0.32±0.03 abc | 0.21±0.03 ab | 18.10±1.76 bcd | 0.15±0.02 e | 0.12±0.02 c | |
| QX13 | 29.83±2.42 abc | 0.26±0.06 bc | 0.21±0.04 ab | 20.43±2.15 abc | 0.19±0.06 abcde | 0.15±0.03 abc | |
表2 根瘤菌与大豆匹配性筛选中植株性状的比较
Table 2 Comparison of plant traits between rhizobium and soybean in matching screening
| Rhizobium | Jinda 88 | Kefeng 1 | |||||
|---|---|---|---|---|---|---|---|
| Plant height/cm | Shoot dry weight/g | Root dry weight/g | Plant height/cm | Shoot dry weight/g | Root dry weight/g | ||
| CK | 26.65±4.78 c | 0.23±0.07 c | 0.19±0.06 abc | 19.55±1.93 abcd | 0.18±0.02 bcde | 0.15±0.01 abc | |
| USDA110 | 31.40±2.91 abc | 0.26±0.04 bc | 0.18±0.02 abc | 19.83±0.45 abcd | 0.20±0.02 abcd | 0.14±0.00 bc | |
| TG1 | 30.15±4.16 abc | 0.28±0.05 abc | 0.19±0.01 abc | 21.80±2.17 a | 0.20±0.03 abc | 0.16±0.02 ab | |
| TG14 | 33.02±3.04 abc | 0.26±0.05 bc | 0.20±0.02 abc | 19.30±1.06 abcd | 0.16±0.03 cde | 0.12±0.02 c | |
| TG20 | 32.10±5.05 abc | 0.25±0.04 c | 0.19±0.02 abc | 16.40±1.67 d | 0.15±0.02 de | 0.12±0.01 c | |
| TG37 | 32.73±3.83 abc | 0.31±0.01 abc | 0.23±0.03 a | 21.43±2.35 ab | 0.23±0.01 a | 0.18±0.02 a | |
| TG53 | 34.45±4.74 abc | 0.28±0.04 abc | 0.21±0.03 abc | 20.37±1.29 abc | 0.17±0.02 bcde | 0.13±0.02 bc | |
| TG79 | 28.68±1.55 bc | 0.26±0.06 bc | 0.19±0.04 abc | 20.13±1.96 abc | 0.18±0.04 bcde | 0.14±0.03 bc | |
| GL11 | 30.63±5.23 abc | 0.36±0.02 a | 0.22±0.02 ab | 17.20±1.57 cd | 0.19±0.01 abcde | 0.14±0.00 bc | |
| GL16 | 31.77±2.96 abc | 0.25±0.03 c | 0.16±0.00 bc | 19.37±0.85 abcd | 0.18±0.01 bcde | 0.12±0.01 c | |
| GL17 | 32.33±5.72 abc | 0.31±0.07 abc | 0.19±0.04 abc | 18.23±2.41 bcd | 0.17±0.01 bcde | 0.12±0.02 c | |
| GL19 | 33.03±1.76 abc | 0.28±0.03 abc | 0.22±0.01 ab | 20.93±0.71 ab | 0.18±0.02 bcde | 0.13±0.01 bc | |
| GL28 | 34.60±2.25 ab | 0.30±0.02 abc | 0.21±0.03 ab | 19.67±1.11 abcd | 0.20±0.02 abc | 0.14±0.02 bc | |
| GL40 | 28.87±8.18 bc | 0.31±0.05 abc | 0.15±0.07 c | 16.97±2.30 cd | 0.16±0.03 cde | 0.13±0.01 c | |
| GL43 | 33.67±4.53 abc | 0.34±0.09 ab | 0.23±0.06 a | 19.47±2.39 abcd | 0.21±0.01 ab | 0.15±0.01 abc | |
| GL67 | 35.53±4.24 ab | 0.29±0.04 abc | 0.21±0.01 ab | 17.93±2.57 bcd | 0.16±0.00 cde | 0.14±0.02 bc | |
| GL75 | 36.97±0.47 a | 0.32±0.03 abc | 0.21±0.03 ab | 18.10±1.76 bcd | 0.15±0.02 e | 0.12±0.02 c | |
| QX13 | 29.83±2.42 abc | 0.26±0.06 bc | 0.21±0.04 ab | 20.43±2.15 abc | 0.19±0.06 abcde | 0.15±0.03 abc | |
| Rhizobium | Jinda 88 | Kefeng 1 | ||||
|---|---|---|---|---|---|---|
| Fresh nodule weight/mg | Dry nodule weight/mg | Nodule number per plant | Fresh nodule weight/mg | Dry nodule weight/mg | Nodule number per plant | |
| USDA110 | 88.15±15.17 ab | 19.18±4.20 ab | 45.83±16.10 a | 46.73±11.90 abc | 10.00±2.14 abc | 22.00±8.54 abcd |
| TG1 | 116.07±34.50 a | 30.02±8.30 a | 43.67±12.50 a | 53.73±34.00 abc | 10.77±6.90 abc | 26.67±26.80 abc |
| TG14 | 101.15±17.40 ab | 23.15±5.20 ab | 36.33±9.29 ab | 68.70±4.25 abc | 17.57±4.70 a | 25.67±16.10 abc |
| TG20 | 87.73±14.20 ab | 18.30±3.44 b | 42.33±5.86 a | 69.77±13.50 ab | 16.33±4.70 ab | 41.67±18.00 a |
| TG37 | 98.13±4.66 ab | 22.43±2.10 ab | 34.67±12.80 ab | 59.47±27.90 abc | 14.43±7.00 abc | 35.33±23.60 abc |
| TG53 | 77.65±30.64 ab | 19.97±5.30 ab | 40.33±7.39 a | 70.83±19.20 a | 15.27±6.00 abc | 28.67±6.11 abc |
| TG79 | 93.38±40.84 ab | 22.40±9.42 ab | 36.50±18.69 ab | 39.80±20.08 bc | 7.80±2.95 c | 12.00±1.00 cd |
| GL11 | 109.53±57.90 ab | 26.20±16.20 ab | 37.33±20.80 ab | 54.17±6.70 abc | 13.03±1.90 abc | 37.67±8.08 ab |
| GL16 | 32.77±14.45 cd | 7.73±3.55 cde | 28.67±6.51 abc | 46.83±7.92 abc | 9.70±2.54 abc | 35.33±9.50 abc |
| GL17 | 84.97±20.66 ab | 19.97±3.00 ab | 46.33±5.86 a | 42.40±12.88 abc | 10.13±3.40 abc | 24.00±12.12 abc |
| GL19 | 81.40±14.60 ab | 17.50±2.01 bc | 36.33±10.70 ab | 41.37±3.35 abc | 7.70±1.51 c | 22.67±6.11 abcd |
| GL28 | 66.27±21.01 bc | 15.13±5.20 bcd | 17.33±1.53 bcd | 44.40±29.99 abc | 11.50±8.92 abc | 17.67±6.81 bcd |
| GL40 | 95.55±39.39 ab | 22.60±7.07 ab | 47.50±24.75 a | 63.20±9.21 abc | 14.27±2.10 abc | 33.67±13.00 abc |
| GL43 | 124.73±39.90 a | 25.13±7.10 ab | 40.67±9.29 a | 53.80±9.36 abc | 12.83±2.50 abc | 21.67±9.87 abcd |
| GL67 | 94.57±11.71 ab | 21.60±3.31 ab | 41.33±6.11 a | 47.47±6.53 abc | 9.37±1.39 abc | 19.00±2.65 abcd |
| GL75 | 106.27±2.25 ab | 24.33±2.20 ab | 43.33±3.06 a | 59.73±11.60 abc | 13.33±2.80 abc | 25.67±13.30 abc |
| QX13 | 29.30±16.95 cd | 6.13±3.11 de | 14.67±9.61 cd | 38.97±1.85 c | 7.83±1.37 c | 27.33±10.00 abc |
表3 根瘤菌与大豆匹配性筛选中根瘤性状的比较
Table 3 Comparison of nodule traits between rhizobium and soybean in matching screening
| Rhizobium | Jinda 88 | Kefeng 1 | ||||
|---|---|---|---|---|---|---|
| Fresh nodule weight/mg | Dry nodule weight/mg | Nodule number per plant | Fresh nodule weight/mg | Dry nodule weight/mg | Nodule number per plant | |
| USDA110 | 88.15±15.17 ab | 19.18±4.20 ab | 45.83±16.10 a | 46.73±11.90 abc | 10.00±2.14 abc | 22.00±8.54 abcd |
| TG1 | 116.07±34.50 a | 30.02±8.30 a | 43.67±12.50 a | 53.73±34.00 abc | 10.77±6.90 abc | 26.67±26.80 abc |
| TG14 | 101.15±17.40 ab | 23.15±5.20 ab | 36.33±9.29 ab | 68.70±4.25 abc | 17.57±4.70 a | 25.67±16.10 abc |
| TG20 | 87.73±14.20 ab | 18.30±3.44 b | 42.33±5.86 a | 69.77±13.50 ab | 16.33±4.70 ab | 41.67±18.00 a |
| TG37 | 98.13±4.66 ab | 22.43±2.10 ab | 34.67±12.80 ab | 59.47±27.90 abc | 14.43±7.00 abc | 35.33±23.60 abc |
| TG53 | 77.65±30.64 ab | 19.97±5.30 ab | 40.33±7.39 a | 70.83±19.20 a | 15.27±6.00 abc | 28.67±6.11 abc |
| TG79 | 93.38±40.84 ab | 22.40±9.42 ab | 36.50±18.69 ab | 39.80±20.08 bc | 7.80±2.95 c | 12.00±1.00 cd |
| GL11 | 109.53±57.90 ab | 26.20±16.20 ab | 37.33±20.80 ab | 54.17±6.70 abc | 13.03±1.90 abc | 37.67±8.08 ab |
| GL16 | 32.77±14.45 cd | 7.73±3.55 cde | 28.67±6.51 abc | 46.83±7.92 abc | 9.70±2.54 abc | 35.33±9.50 abc |
| GL17 | 84.97±20.66 ab | 19.97±3.00 ab | 46.33±5.86 a | 42.40±12.88 abc | 10.13±3.40 abc | 24.00±12.12 abc |
| GL19 | 81.40±14.60 ab | 17.50±2.01 bc | 36.33±10.70 ab | 41.37±3.35 abc | 7.70±1.51 c | 22.67±6.11 abcd |
| GL28 | 66.27±21.01 bc | 15.13±5.20 bcd | 17.33±1.53 bcd | 44.40±29.99 abc | 11.50±8.92 abc | 17.67±6.81 bcd |
| GL40 | 95.55±39.39 ab | 22.60±7.07 ab | 47.50±24.75 a | 63.20±9.21 abc | 14.27±2.10 abc | 33.67±13.00 abc |
| GL43 | 124.73±39.90 a | 25.13±7.10 ab | 40.67±9.29 a | 53.80±9.36 abc | 12.83±2.50 abc | 21.67±9.87 abcd |
| GL67 | 94.57±11.71 ab | 21.60±3.31 ab | 41.33±6.11 a | 47.47±6.53 abc | 9.37±1.39 abc | 19.00±2.65 abcd |
| GL75 | 106.27±2.25 ab | 24.33±2.20 ab | 43.33±3.06 a | 59.73±11.60 abc | 13.33±2.80 abc | 25.67±13.30 abc |
| QX13 | 29.30±16.95 cd | 6.13±3.11 de | 14.67±9.61 cd | 38.97±1.85 c | 7.83±1.37 c | 27.33±10.00 abc |
| [1] |
Popp C, Ott T. Regulation of signal transduction and bacterial infection during root nodule symbiosis[J]. Curr Opin Plant Biol, 2011, 14(4):458-467.
doi: 10.1016/j.pbi.2011.03.016 URL |
| [2] | 柏宇, 关大伟, 李力, 等. 耐高氮优良大豆根瘤菌株的筛选与鉴定[J]. 大豆科学, 2014, 33(6):861-864. |
| Bai Y, Guan DW, Li L, et al. Screening and characterization of superior nitrogen-tolerance soybean rhizobia[J]. Soybean Sci, 2014, 33(6):861-864. | |
| [3] |
Zhang XX, Guo HJ, Jiao J, et al. Pyrosequencing of rpoB uncovers a significant biogeographical pattern of rhizobial species in soybean rhizosphere[J]. J Biogeogr, 2017, 44(7):1491-1499.
doi: 10.1111/jbi.2017.44.issue-7 URL |
| [4] | 陈文峰. 根瘤菌系统学研究进展与展望[J]. 微生物学通报, 2016, 43(5):1095-1100. |
| Chen WF. Progress and perspective of systematics of rhizobia[J]. Microbiol China, 2016, 43(5):1095-1100. | |
| [5] | 程涛, 焦如珍, 高俊莲. 对两个分离自海南尖峰岭的木本豆科植物根瘤菌菌株的多相鉴定[J]. 林业科学研究, 2015, 28(3):380-386. |
| Cheng T, Jiao RZ, Gao JL. Polyphasic identification of two Rhizobium strains from woody legumes in Jianfengling of Hainan Province[J]. For Res, 2015, 28(3):380-386. | |
| [6] |
Mohammed M, Jaiswal SK, Dakora FD. Distribution and correlation between phylogeny and functional traits of cowpea(Vigna unguiculata L. Walp. )-nodulating microsymbionts from Ghana and South Africa[J]. Sci Rep, 2018, 8:18006.
doi: 10.1038/s41598-018-36324-0 URL |
| [7] |
Zhang YM, Tian CF, Sui XH, et al. Robust markers reflecting phylogeny and taxonomy of rhizobia[J]. PLoS One, 2012, 7(9):e44936.
doi: 10.1371/journal.pone.0044936 URL |
| [8] | 蒲强, 谭志远, 彭桂香, 等. 根瘤菌分类的新进展[J]. 微生物学通报, 2016, 43(3):619-633. |
| Pu Q, Tan ZY, Peng GX, et al. Advances in rhizobia taxonomy[J]. Microbiol China, 2016, 43(3):619-633. | |
| [9] |
Borba MP, Ballarini AE, Witusk JPD, et al. Evaluation of BOX-PCR and REP-PCR as molecular typing tools for Antarctic Streptomyces[J]. Curr Microbiol, 2020, 77(11):3573-3581.
doi: 10.1007/s00284-020-02199-6 URL |
| [10] | 陈文新, 陈文峰. 发挥生物固氮作用减少化学氮肥用量[J]. 中国农业科技导报, 2004, 6(6):3-6. |
| Chen WX, Chen WF. Exertion of biological nitrogen fixation in order to reducing the consumption of chemical nitrogenous fertilizer[J]. Rev China Agric Sci Technol, 2004, 6(6):3-6. | |
| [11] | 王金生, 丁宁, 吴俊江, 等. 大豆根瘤菌接种效应及竞争结瘤能力分析[J]. 大豆科学, 2017, 36(5):761-767. |
| Wang JS, Ding N, Wu JJ, et al. Analysis of the inoculation effect of soybean rhizobia and the competitive nodulation ability[J]. Soybean Sci, 2017, 36(5):761-767. | |
| [12] |
Fan KK, Delgado-Baquerizo M, Guo XS, et al. Suppressed N fixation and diazotrophs after four decades of fertilization[J]. Microbiome, 2019, 7(1):143.
doi: 10.1186/s40168-019-0757-8 URL |
| [13] | 高俊莲, 孙建光, 陈文新. 斜茎黄芪根瘤菌结瘤基因nodA PCR扩增及PCR-RFLP分析[J]. 微生物学杂志, 2006, 26(4):1-5. |
| Gao JL, Sun JG, Chen WX. PCR amplification and PCR-RFLP analysis of nodulation gene nodA of rhizobia from Astragalus adsurgens[J]. J Microbiol, 2006, 26(4):1-5. | |
| [14] |
Fredriksson NJ, Hermansson M, Wilén BM. The choice of PCR primers has great impact on assessments of bacterial community diversity and dynamics in a wastewater treatment plant[J]. PLoS One, 2013, 8(10):e76431.
doi: 10.1371/journal.pone.0076431 URL |
| [15] |
Wang JY, Wang R, Zhang YM, et al. Bradyrhizobium daqingense sp. nov., isolated from soybean nodules[J]. Int J Syst Evol Microbiol, 2013, 63(Pt 2):616-624.
doi: 10.1099/ijs.0.034280-0 URL |
| [16] |
Zhang YM, Li Y, Chen WF, et al. Biodiversity and biogeography of rhizobia associated with soybean plants grown in the North China plain[J]. Appl Environ Microbiol, 2011, 77(18):6331-6342.
doi: 10.1128/AEM.00542-11 URL |
| [17] | 葛诚, 徐玲玫, 樊惠. 超慢型大豆根瘤菌的分离和初步鉴定[J]. 中国农业科学, 1987, 20(4):96. |
| Ge C, Xu LM, Fan H. Isolated and initially identified on extra slow-growing soybean rhizobia[J]. Sci Agric Sin, 1987, 20(4):96. | |
| [18] |
Han LL, Wang ET, Han TX, et al. Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang, China[J]. Plant Soil, 2009, 324(1/2):291-305.
doi: 10.1007/s11104-009-9956-6 URL |
| [19] | 张璐. 不同品种大豆与土壤中根瘤菌的多样性研究[D]. 杨凌:西北农林科技大学, 2017. |
| Zhang L. Diversity of Rhizobium from the roots of different soybeans varieties and soil[D]. Yangling:Northwest A & F University, 2017. | |
| [20] | 史清亮, 白成云, 温月香, 等. 山西省费氏中华根瘤菌数量分布及特性探析[J]. 中国生态农业学报, 2007, 15(3):120-122. |
| Shi QL, Bai CY, Wen YX, et al. Quantitative distribution and character analysis of Sinorchizobium fredii in Shanxi province[J]. Chin J Eco Agric, 2007, 15(3):120-122. Chinese Journal of Eco-Agriculture, 2007(3):120-122. | |
| [21] | 张红侠. 黄土高原地区大豆根瘤菌遗传多样性、高效菌株筛选及应用效果的研究[D]. 北京:中国农业科学院, 2010. |
| Zhang HX. Genetic diversity, screening and application effect of soybean rhizobia in Loess Plateau[D]. Beijing:Chinese Academy of Agricultural Sciences, 2010. | |
| [22] | 杨凤环, 李正楠, 姬惜珠, 等. BOX-PCR技术在微生物多样性研究中的应用[J]. 微生物学通报, 2008, 35(8):1282-1286. |
| Yang FH, Li ZN, Ji XZ, et al. Applications of BOX-PCR technique in diversity study of microorganisms[J]. Microbiology, 2008, 35(8):1282-1286. | |
| [23] |
Loureiro MDF, Kaschuk G, Alberton O, et al. Soybean[Glycine max(L.)Merrill]rhizobial diversity in Brazilian oxisols under various soil, cropping, and inoculation managements[J]. Biol Fertil Soils, 2007, 43(6):665-674.
doi: 10.1007/s00374-006-0146-x URL |
| [24] | 王卫卫, 关大伟, 马鸣超, 等. 东北地区大豆根瘤菌遗传多样性与系统发育研究[J]. 大豆科学, 2013, 32(4):433-437. |
| Wang WW, Guan DW, Ma MC, et al. Genetic diversity and phylogeny of soybean rhizobia isolated from northeast China[J]. Soybean Sci, 2013, 32(4):433-437. | |
| [25] | 张红侠, 冯瑞华, 关大伟, 等. 黄土高原地区优良大豆根瘤菌的筛选与接种方式研究[J]. 大豆科学, 2010, 29(6):996-1002. |
| Zhang HX, Feng RH, Guan DW, et al. Screening of superior soybean rhizobial strains and analyzing of different inoculation methods in loess plateau region of China[J]. Soybean Sci, 2010, 29(6):996-1002. | |
| [26] | 蒋攀. 四川大豆根瘤菌高效菌株的初步筛选[D]. 雅安:四川农业大学, 2013. |
| Jiang P. Preliminary screening of high efficient soybean rhizobia strains in Sichuan Province[D]. Ya’an:Sichuan Agricultural University, 2013. | |
| [27] | 伍惠, 钟喆栋, 樊伟, 等. 8株优良大豆根瘤菌与不同地区27个大豆主栽品种的匹配性研究[J]. 大豆科学, 2017, 36(3):405-418. |
| Wu H, Zhong ZD, Fan W, et al. Symbiotic compatibility among eight elite soybean rhizobia strains and twenty-seven soybean cultivars from different planting regions[J]. Soybean Sci, 2017, 36(3):405-418. |
| [1] | 方澜, 黎妍妍, 江健伟, 成胜, 孙正祥, 周燚. 盘龙参内生真菌胞内细菌7-2H的分离鉴定和促生特性研究[J]. 生物技术通报, 2023, 39(8): 272-282. |
| [2] | 颜珲璘, 芦光新, 邓晔, 顾松松, 颜程良, 马坤, 赵阳安, 张海娟, 王英成, 周学丽, 窦声云. 高寒地区根瘤菌拌种对禾/豆混播土壤微生物群落的影响[J]. 生物技术通报, 2022, 38(10): 204-215. |
| [3] | 焦健, 刘克寒, 田长富. 根瘤菌铁转运代谢及其调控机制研究进展[J]. 生物技术通报, 2019, 35(10): 7-17. |
| [4] | 陈雪莲, 江高飞, 钟增涛. 基因水平转移在根瘤菌进化中的研究进展[J]. 生物技术通报, 2019, 35(10): 18-24. |
| [5] | 董汝, 曹扬荣. 豆科植物-根瘤菌共生固氮的免疫调控机制[J]. 生物技术通报, 2019, 35(10): 25-33. |
| [6] | 林丽, 李杨瑞, 安千里. 甘蔗联合固氮的回顾与展望[J]. 生物技术通报, 2019, 35(10): 46-56. |
| [7] | 彭卫福, 吴志明, 陈未, 曾勇军, 李昆太. 拮抗多种植物病原真菌Streptomyces triostinicus C2的分离与鉴定[J]. 生物技术通报, 2016, 32(7): 106-111. |
| [8] | 刘毅, 姚粟, 李辉, 刘洋, 刘勇, 刘波, 程池. 三种DNA指纹图谱技术在菌株分型中的应用[J]. 生物技术通报, 2015, 31(6): 81-86. |
| [9] | 李正, 单辉辉, 齐雅琳, 刘磊, 韩素贞. 甘肃酒泉等地区豆科植物根瘤菌的遗传多样性和系统发育分析[J]. 生物技术通报, 2014, 0(10): 188-195. |
| [10] | 李珊珊, 李海英, 于冰. 导入nodD-lba串联基因的根瘤菌工程菌株的构建[J]. 生物技术通报, 2013, 0(8): 119-123. |
| [11] | 单辉辉 李正 韩素贞. 两株藏南地区土壤根瘤菌分类地位的确定[J]. 生物技术通报, 2013, 0(4): 158-166. |
| [12] | 于海鹏;潘钰;蒙明明;李海英;. 大豆血红蛋白基因lba转化根瘤菌工程菌株的构建[J]. , 2012, 0(02): 69-74. |
| [13] | 张伟涛;程国军;周俊初;. 利用TAIL-PCR克隆耐盐基因及其分析[J]. , 2010, 0(11): 166-170. |
| [14] | 高俊莲;孙建光;陈文新;. 斜茎黄芪根瘤菌谷氨酰胺合成酶基因多样性研究[J]. , 2007, 0(01): 124-127. |
| [15] | . 文摘[J]. , 2000, 0(05): 56-59. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||