








生物技术通报 ›› 2023, Vol. 39 ›› Issue (3): 218-231.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0802
王涛1,2( ), 漆思雨1,2, 韦朝领3, 王艺清1,2, 戴浩民1,2, 周喆1,2, 曹士先4, 曾雯1,2(
), 漆思雨1,2, 韦朝领3, 王艺清1,2, 戴浩民1,2, 周喆1,2, 曹士先4, 曾雯1,2( ), 孙威江1,2(
), 孙威江1,2( )
)
收稿日期:2022-06-29
出版日期:2023-03-26
发布日期:2023-04-10
通讯作者:
曾雯,女,博士,讲师,研究方向:茶树栽培育种与生物技术;E-mail: wenzeng_t@163.com;作者简介:王涛,男,硕士研究生,研究方向:茶树栽培育种与生物技术;E-mail: wangtwtao0827@163.com
基金资助:
WANG Tao1,2( ), QI Si-yu1,2, WEI Chao-ling3, WANG Yi-qing1,2, DAI Hao-min1,2, ZHOU Zhe1,2, CAO Shi-xian4, ZENG Wen1,2(
), QI Si-yu1,2, WEI Chao-ling3, WANG Yi-qing1,2, DAI Hao-min1,2, ZHOU Zhe1,2, CAO Shi-xian4, ZENG Wen1,2( ), SUN Wei-jiang1,2(
), SUN Wei-jiang1,2( )
)
Received:2022-06-29
Published:2023-03-26
Online:2023-04-10
摘要:
本课题组前期通过转录组筛选出2个与‘白鸡冠’茶树(Camellia sinensis)叶片白化相关的基因(CSS0013384和CSS0036305),为探明CSS0013384和CSS0036305在白化茶树中的表达模式与其互作蛋白,以‘白鸡冠’茶树叶片为材料,克隆CSS0013384和CSS0036305 cDNA全长序列,利用生物信息学、酵母单杂交和酵母双杂交,分析其蛋白理化性质、系统进化树、染色体定位、基因结构、蛋白结构、蛋白调控与互作网络和基因表达模式。生物信息学分析结果表明,CSS0013384和CSS0036305分别属于三角状五肽蛋白(pentatricopeptide repeat protein, PPR)和伴侣蛋白(chaperone, CPN60-like)家族,其蛋白质编码区(coding sequence, CDS)长度为1 893 bp和1 752 bp,编码氨基酸个数为631和575,蛋白质质量为71.87 kD和60.79 kD,等电点为8.93和6.21。亚细胞定位预测结果表明,CSS0013384定位于叶绿体,CSS0036305定位于线粒体。通过白鸡冠茶树第二叶的遮阴和恢复光照处理与不同叶色茶树品种的RT-qPCR发现,CSS0013384和CSS0036305在白化芽叶中高表达。CSS0002807属于PIF转录因子家族,酵母单杂交结果表明CSS0002807可以结合CSS0013384启动子。CSS0013384和CSS0036305在白化茶树叶片中可能参与叶绿体和线粒体发育,在叶片白化过程中发挥重要作用,结果可为进一步探究茶树叶片白化机理提供参考。
王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231.
WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis)[J]. Biotechnology Bulletin, 2023, 39(3): 218-231.
| 基因名称 Gene name | 上游引物 Sense primer(5'-3') | 下游引物 Anti-sense primer(5'-3') | 备注 Note |
|---|---|---|---|
| CSS0013384 | ATGGCCATCCCCAGTTCTCCAGACTGGTCTGTG | TTACATGGTGCCAAAGCTCTGTTGCAACCATTTTGC | 基因克隆引物 |
| CSS0036305 | ATGTACAGAGTTGGTGCAGCCTTCGCTTCATCGAT | TCAGTAACCCATATCATCCAAATTTGGCATTCGAT | 基因克隆引物 |
| qCSS0013384 | TGGGGAAAACAGAAGGGCTAA | CAACACCACCACCTGGAACAA | 荧光定量引物 |
| qCSS0036305 | CCAAAATGAAGGAAAGAGGGTG | TCAAAGCGGAGCGATAGAGC | 荧光定量引物 |
| β-Actin | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT | RNA验证 |
| pAbAi- CSS0013384pro | AAATGATGAATTGAAAAGCTTAAGAAATTTTTTGTTGAG | ATACAGAGCACATGCCTCGAGAATTTTGCCTTGCATCAT | 酵母单杂交 |
| AD-CSS0002807 | GCCATGGAGGCCAGTGAATTCATGAAAGGAATCATGAA | CTCGATGGATCTCACTTACTTGAGCTAGAA | 酵母单杂交 |
| BK-CSS0003881 | ATGGCCATGGAGGCCGAATTCATGGCATCAACTTTGGCT | CCGCTGCAGGTCGACGGATCTTAGTAACCGTATCCTGA | 酵母双杂交 |
| BK-CSS0041773 | ATGGCCATGGAGGCCGAATTCATGGCATCAACTTTGGCT | CCGCTGCAGGTCGACGGATCTTAGTAACCGTATCCTGA | 酵母双杂交 |
| BK-CSS0043610 | ATGGCCATGGAGGCCGAATTCATGTACCGCTTCGCAG | CCGCTGCAGGTCGACGGATCTCAAAAATCCATACCACC | 酵母双杂交 |
| AD-CSS0036305 | GCCATGGAGGCCAGTGAATTCATGTACAGAGTTGGTGC | CAGCTCGAGCTCGATGGATCTCAGTAACCCATATCATCC | 酵母双杂交 |
| BK-CSS0036305 | ATGGCCATGGAGGCCGAATTCATGTACAGAGTTGGTGCA | CCGCTGCAGGTCGACGGATCTCAGTAACCCATATCATCC | 酵母双杂交 |
| AD-CSS0003881 | GCCATGGAGGCCAGTGAATTCATGGCATCAACTTTGGCT | CAGCTCGAGCTCGATGGATCTTAGTAACCGTATCCTGA | 酵母双杂交 |
| AD-CSS0041773 | GCCATGGAGGCCAGTGAATTCATGGCATCAACTTTGGCT | CAGCTCGAGCTCGATGGATCTTAGTAACCGTATCCTGA | 酵母双杂交 |
| AD-CSS0043610 | GCCATGGAGGCCAGTGAATTCATGTACCGCTTCGCAG | CAGCTCGAGCTCGATGGATCTCAAAAATCCATACCAC | 酵母双杂交 |
表1 试验所需引物
Table 1 Primers required for the experiment
| 基因名称 Gene name | 上游引物 Sense primer(5'-3') | 下游引物 Anti-sense primer(5'-3') | 备注 Note |
|---|---|---|---|
| CSS0013384 | ATGGCCATCCCCAGTTCTCCAGACTGGTCTGTG | TTACATGGTGCCAAAGCTCTGTTGCAACCATTTTGC | 基因克隆引物 |
| CSS0036305 | ATGTACAGAGTTGGTGCAGCCTTCGCTTCATCGAT | TCAGTAACCCATATCATCCAAATTTGGCATTCGAT | 基因克隆引物 |
| qCSS0013384 | TGGGGAAAACAGAAGGGCTAA | CAACACCACCACCTGGAACAA | 荧光定量引物 |
| qCSS0036305 | CCAAAATGAAGGAAAGAGGGTG | TCAAAGCGGAGCGATAGAGC | 荧光定量引物 |
| β-Actin | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT | RNA验证 |
| pAbAi- CSS0013384pro | AAATGATGAATTGAAAAGCTTAAGAAATTTTTTGTTGAG | ATACAGAGCACATGCCTCGAGAATTTTGCCTTGCATCAT | 酵母单杂交 |
| AD-CSS0002807 | GCCATGGAGGCCAGTGAATTCATGAAAGGAATCATGAA | CTCGATGGATCTCACTTACTTGAGCTAGAA | 酵母单杂交 |
| BK-CSS0003881 | ATGGCCATGGAGGCCGAATTCATGGCATCAACTTTGGCT | CCGCTGCAGGTCGACGGATCTTAGTAACCGTATCCTGA | 酵母双杂交 |
| BK-CSS0041773 | ATGGCCATGGAGGCCGAATTCATGGCATCAACTTTGGCT | CCGCTGCAGGTCGACGGATCTTAGTAACCGTATCCTGA | 酵母双杂交 |
| BK-CSS0043610 | ATGGCCATGGAGGCCGAATTCATGTACCGCTTCGCAG | CCGCTGCAGGTCGACGGATCTCAAAAATCCATACCACC | 酵母双杂交 |
| AD-CSS0036305 | GCCATGGAGGCCAGTGAATTCATGTACAGAGTTGGTGC | CAGCTCGAGCTCGATGGATCTCAGTAACCCATATCATCC | 酵母双杂交 |
| BK-CSS0036305 | ATGGCCATGGAGGCCGAATTCATGTACAGAGTTGGTGCA | CCGCTGCAGGTCGACGGATCTCAGTAACCCATATCATCC | 酵母双杂交 |
| AD-CSS0003881 | GCCATGGAGGCCAGTGAATTCATGGCATCAACTTTGGCT | CAGCTCGAGCTCGATGGATCTTAGTAACCGTATCCTGA | 酵母双杂交 |
| AD-CSS0041773 | GCCATGGAGGCCAGTGAATTCATGGCATCAACTTTGGCT | CAGCTCGAGCTCGATGGATCTTAGTAACCGTATCCTGA | 酵母双杂交 |
| AD-CSS0043610 | GCCATGGAGGCCAGTGAATTCATGTACCGCTTCGCAG | CAGCTCGAGCTCGATGGATCTCAAAAATCCATACCAC | 酵母双杂交 |
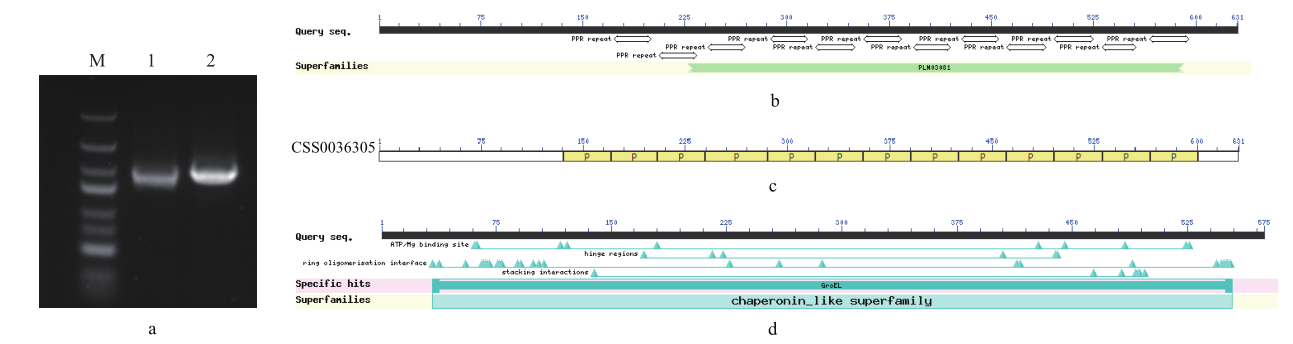
图1 基因扩增电泳图及蛋白结构预测 a:茶树CSS0013384和CSS0036305 PCR扩增电泳图(M:marker;1:CSS0036305;2:CSS0013384);b:茶树CSS0013384蛋白保守结构域预测;c:CSS0013384 motif分布图;d:茶树CSS0036305蛋白保守结构域预测
Fig. 1 Gene amplification electropherogram and protein structure prediction a: Electrophoresis of PCR amplification of CSS0013384 and CSS0036305 in C. sinensis. b: Conserved domain prediction of CSS0013384 in C. sinensis. c: Motif distribution map of CSS0013384. d: Conserved domain prediction of CSS0036305 in C. sinensis
| 基因 Gene | 分子量 Mw/kD | 等电点 pI | 不稳定系数 Instability index | 脂肪族氨基酸指数Aliphatic index | 平均疏水性 GRAVY | 信号肽 Signal peptide | 跨膜结构 Transmembrane domain | 亚细胞定位预测 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| CSS0013384 | 71.87 | 8.93 | 40.73 | 85.90 | -0.167 | 无 | 无 | 叶绿体 |
| CSS0036305 | 60.79 | 6.21 | 21.85 | 103.99 | 0.002 | 无 | 无 | 线粒体 |
表2 CSS0013384和CSS0036305蛋白的基本理化性质
Table 2 Basic physicochemical properties of CSS0013384 and CSS0036305 protein
| 基因 Gene | 分子量 Mw/kD | 等电点 pI | 不稳定系数 Instability index | 脂肪族氨基酸指数Aliphatic index | 平均疏水性 GRAVY | 信号肽 Signal peptide | 跨膜结构 Transmembrane domain | 亚细胞定位预测 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| CSS0013384 | 71.87 | 8.93 | 40.73 | 85.90 | -0.167 | 无 | 无 | 叶绿体 |
| CSS0036305 | 60.79 | 6.21 | 21.85 | 103.99 | 0.002 | 无 | 无 | 线粒体 |
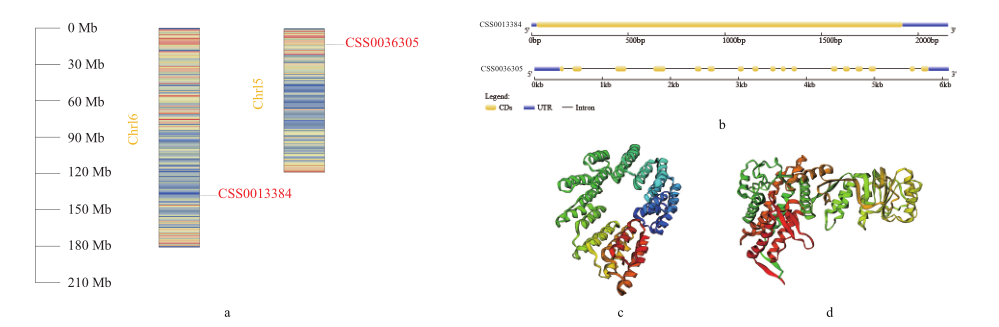
图2 基因染色体定位、基因结构及蛋白三级结构 a:茶树CSS0013384和CSS0036305染色体定位;b:茶树CSS0013384和CSS0036305基因结构;c:茶树CSS0013384蛋白三级结构预测;d:茶树CSS0036305蛋白三级结构预测
Fig. 2 Gene chromosomal location, gene structure and protein tertiary structure a: Chromosomal localization of CSS0013384 and CSS0036305 in tea plant(C. sinensis). b: Gene structure of CSS0013384 and CSS0036305 in tea plant. c: The tertiary structure prediction of CSS0013384 in tea plant. d: The tertiary structure prediction of CSS0036305 in tea plant

图3 基因进化树 a:茶树CSS0013384与不同植物的PPR的进化树分析;b:茶树CSS0036305与不同植物的CPN60-like的进化树分析
Fig. 3 Genetic tree a: PPR neighbor-joining tree of CSS0036305 in tea pant and other plants. b: CPN60-like neighbor-joining tree of CSS0036305 in tea plant and other plants
| 类型Type | 基因名称Gene name | 注释Function | |
|---|---|---|---|
| CSS0013384 | CSS0036305 | ||
| 光响应 Light response | A-box(2)、ATCT-motif(1)、Box 4(6)、Box Ⅱ(1)、GA-motif(1)、GATA-motif(1)、G-box(5)、I-box(1) | Box 4(1)、Box Ⅱ(1)、GATA-motif(3)、G-box(2)、GT1-motif(2)、TCCC-motif(1)、TCT-motif(2)、 | 参与光反应的顺式作用元件cis-acting element involved in light responsiveness |
| 转录因子结合位点 Transcription factor binding site | MRE(1)、MBS(1)、MBSI(1)、Myb-binding site(1) | HD-Zip3(1)、MRE(1)、MBS(2) | 转录因子结合位点 Transcription factor binding site |
| 激素响应 Hormone response | AAGAA-motif(1)、ABRE(2)、ABRE2(1) | — | 参与脱落酸反应的顺式作用元件cis-acting element involved in the abscisic acid responsiveness |
| ERE(3) | — | 参与乙烯反应的顺式作用元件 cis-acting element involved in ethylene responsiveness | |
| TCA-element(1) | TCA-element(2) | 与水杨酸反应有关的顺式作用元件 cis-acting element involved in salicylic acid responsiveness | |
| — | as-1(3)、CGTCA-motif(3)、TGACG-motif(3) | 参与MeJA反应的顺式作用元件 cis-acting element involved in MeJA responsiveness | |
| 组织特异性 Tissue specificity | CAT-box(2)、CCGTCC motif(4)、GCN4 motif(1) | — | 组织特异性元件 Tissue specific element |
| 胁迫响应 Stress response | ARE(1)、STRE(1)、WUN-motif(4) | ACE(1)、ARE(4)、STRE(2)、WRE3(2)、WUN-motif(2) | 胁迫响应元件Stress response element |
表3 Prediction of cis-elements in the upstream region of CSS0013384和CSS0036305 in tea plant
Table 3
| 类型Type | 基因名称Gene name | 注释Function | |
|---|---|---|---|
| CSS0013384 | CSS0036305 | ||
| 光响应 Light response | A-box(2)、ATCT-motif(1)、Box 4(6)、Box Ⅱ(1)、GA-motif(1)、GATA-motif(1)、G-box(5)、I-box(1) | Box 4(1)、Box Ⅱ(1)、GATA-motif(3)、G-box(2)、GT1-motif(2)、TCCC-motif(1)、TCT-motif(2)、 | 参与光反应的顺式作用元件cis-acting element involved in light responsiveness |
| 转录因子结合位点 Transcription factor binding site | MRE(1)、MBS(1)、MBSI(1)、Myb-binding site(1) | HD-Zip3(1)、MRE(1)、MBS(2) | 转录因子结合位点 Transcription factor binding site |
| 激素响应 Hormone response | AAGAA-motif(1)、ABRE(2)、ABRE2(1) | — | 参与脱落酸反应的顺式作用元件cis-acting element involved in the abscisic acid responsiveness |
| ERE(3) | — | 参与乙烯反应的顺式作用元件 cis-acting element involved in ethylene responsiveness | |
| TCA-element(1) | TCA-element(2) | 与水杨酸反应有关的顺式作用元件 cis-acting element involved in salicylic acid responsiveness | |
| — | as-1(3)、CGTCA-motif(3)、TGACG-motif(3) | 参与MeJA反应的顺式作用元件 cis-acting element involved in MeJA responsiveness | |
| 组织特异性 Tissue specificity | CAT-box(2)、CCGTCC motif(4)、GCN4 motif(1) | — | 组织特异性元件 Tissue specific element |
| 胁迫响应 Stress response | ARE(1)、STRE(1)、WUN-motif(4) | ACE(1)、ARE(4)、STRE(2)、WRE3(2)、WUN-motif(2) | 胁迫响应元件Stress response element |
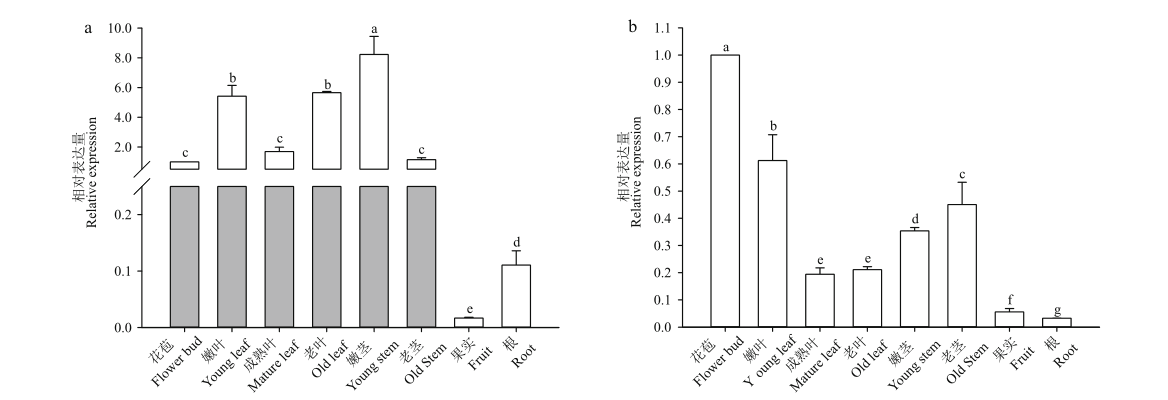
图4 茶树不同部位基因的表达模式 a:CSS0013384在茶树不同组织部位表达模式;b:CSS0036305在茶树不同组织部位表达模式。不同小写字母表示在P<0.05水平差异显著
Fig. 4 Expression patterns of genes in different parts of tea plant a: Expression pattern of CSS0013384 in different tissues of tea plant. b: Expression pattern of CSS0036305 in different tissues of tea plant. Different lower letters mean the significant difference in P<0.05
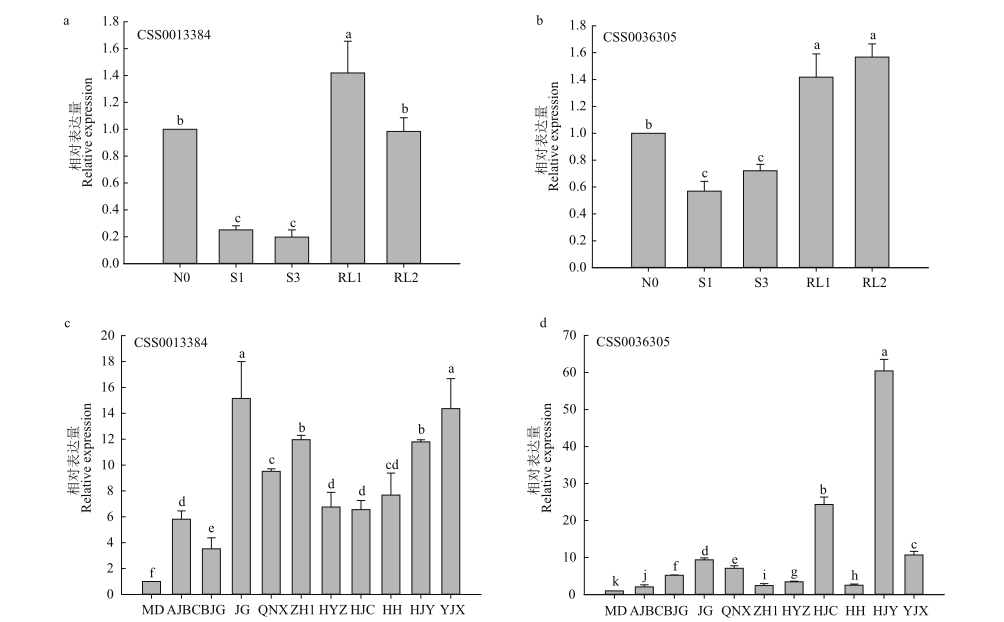
图5 遮阴和恢复光照与不同茶树品种间基因表达模式 a:茶树CSS0013384在遮阴和恢复光照下的表达模式分析;b:茶树CSS0036305在遮阴和恢复光照下的表达模式分析;c:茶树CSS0013384在不同茶树品种中的表达分析;d:茶树CSS0036305在不同茶树品种中的表达分析。N0:不遮阴;S1:遮阴1 d;S3:遮阴3 d;RL1:恢复光照1 d;RL2:恢复光照2 d;MD:‘金牡丹’;AJBC:‘安吉白茶’;BJG:‘白鸡冠’;JG:‘金光’;QNX:‘千年雪’;ZH1:‘中黄1号’;HYZ:‘黄芽早’;HJC:‘黄金茶’;HH:黄化品系;HJY:‘黄金芽’;YJX:‘御金香’。不同字母表示样本间存在显著差异(P<0.05);遮阴和恢复光照处理下基因的相对表达量以N0为对照,不同茶树品种间的基因表达模式以‘金牡丹’(MD)为对照
Fig. 5 Gene expression patterns of genes from different tea cultivars under shading and restored light a: Expression pattern analysis of tea plant CSS0013384 under shading and restored light. b: Expression pattern analysis of tea plant CSS0036305 under shading and restored light. c: Expression analysis of CSS0013384 in different tea plant varieties. d: Expression analysis of CSS0036305 in different tea plant varieties. N0: No shading. S1: Shading 1 d. S3: Shading 3 d. RL1: Restored light for 1 d. RL2: Restored light for 2 d. MD: Jinmudan. AJBC: Anjibaiha. BJG: Baijiguan. JG: Jinguang. QNX: Qiannianxue. ZH1: Zhonghuang 1. HYZ: Huangyazao. HJC: Huangjincha. HH: Albino tea plant germplasm. HJY: Huangjinya. YJX: Yujinxiang. Different letters indicate significant differences between samples(P<0.05). Relative expressions of genes under shading and restored light were compared with N0, and the gene expression patterns among different tea varieties were compared with Jinmudan(MD)
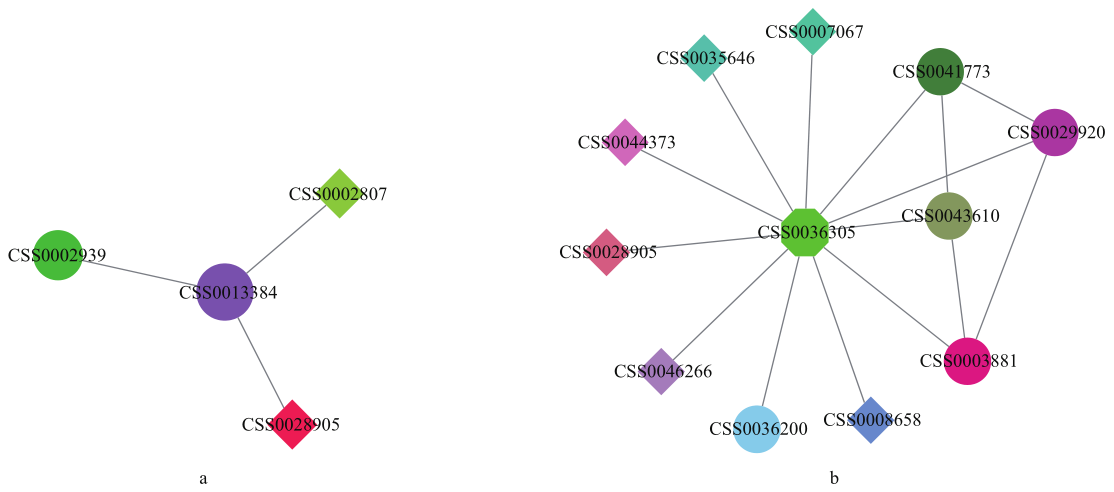
图6 基因互作网络预测图 a:茶树CSS0013384蛋白互作网络;b:茶树CSS0036305蛋白互作网络。菱形代表转录因子,圆形代表功能基因
Fig. 6 Gene interaction network prediction map a: Protein interaction network of CSS0013384 in tea plant. b: Protein interaction network of CSS0036305 in tea plant. Diamond represents transcription factor. Circle represents functional gene
| 靶蛋白 Target protein | 互作蛋白 Interacting protein | 基因注释 Gene annotation |
|---|---|---|
| CSS0036305 | CSS0029920 | TCP-1/cpn60 伴侣蛋白家族蛋白 TCP-1/cpn60 chaperonin family protein |
| CSS0043610 | TCP-1/cpn60 伴侣蛋白家族蛋白 TCP-1/cpn60 chaperonin family protein | |
| CSS0003881 | β Rubisco大亚基结合蛋白亚基 Rubisco large subunit-binding protein subunit beta | |
| CSS0041773 | β Rubisco大亚基结合蛋白亚基 Rubisco large subunit-binding protein subunit beta | |
| CSS0036200 | Rubisco亚基结合蛋白β亚基 Rubisco subunit binding-protein beta subunit | |
| CSS0044373 | GAGA结合转录激活因子 GAGA-binding transcriptional activator | |
| CSS0007067 | GAGA结合转录激活因子 GAGA-binding transcriptional activator | |
| CSS0008658 | GAGA结合转录激活因子 GAGA-binding transcriptional activator | |
| CSS0046266 | MYB_related蛋白 MYB_related protein | |
| CSS0035646 | MYB_related蛋白 MYB_related protein | |
| CSS0028905 | MADS-box蛋白 MADS-box protein | |
| CSS0013384 | CSS0002939 | 鸟嘌呤核苷酸结合蛋白亚基β-蛋白异构体X2 Guanine nucleotide-binding protein subunit beta-like protein isoform X2 |
| CSS0002807 | PIF转录因子 Transcription factor PIF7 | |
| CSS0028905 | MADS家族蛋白 MADS-box protein |
表4 CSS0013384和CSS0036305互作蛋白功能注释
Table 4 Functional annotations of interacting proteins of CSS0013384 and CSS0036305
| 靶蛋白 Target protein | 互作蛋白 Interacting protein | 基因注释 Gene annotation |
|---|---|---|
| CSS0036305 | CSS0029920 | TCP-1/cpn60 伴侣蛋白家族蛋白 TCP-1/cpn60 chaperonin family protein |
| CSS0043610 | TCP-1/cpn60 伴侣蛋白家族蛋白 TCP-1/cpn60 chaperonin family protein | |
| CSS0003881 | β Rubisco大亚基结合蛋白亚基 Rubisco large subunit-binding protein subunit beta | |
| CSS0041773 | β Rubisco大亚基结合蛋白亚基 Rubisco large subunit-binding protein subunit beta | |
| CSS0036200 | Rubisco亚基结合蛋白β亚基 Rubisco subunit binding-protein beta subunit | |
| CSS0044373 | GAGA结合转录激活因子 GAGA-binding transcriptional activator | |
| CSS0007067 | GAGA结合转录激活因子 GAGA-binding transcriptional activator | |
| CSS0008658 | GAGA结合转录激活因子 GAGA-binding transcriptional activator | |
| CSS0046266 | MYB_related蛋白 MYB_related protein | |
| CSS0035646 | MYB_related蛋白 MYB_related protein | |
| CSS0028905 | MADS-box蛋白 MADS-box protein | |
| CSS0013384 | CSS0002939 | 鸟嘌呤核苷酸结合蛋白亚基β-蛋白异构体X2 Guanine nucleotide-binding protein subunit beta-like protein isoform X2 |
| CSS0002807 | PIF转录因子 Transcription factor PIF7 | |
| CSS0028905 | MADS家族蛋白 MADS-box protein |
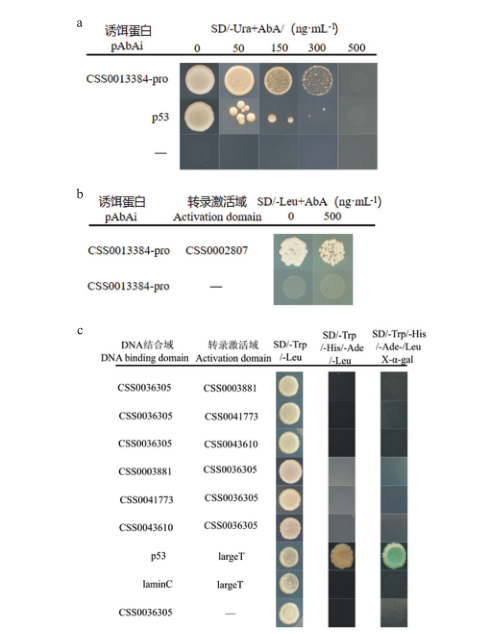
图7 酵母单杂交和酵母双杂交结果图 a:CSS0013384启动子自激活检测;b:CSS0013384酵母单杂交结果;c:CSS0036305酵母双杂交结果。SD/-Ura+AbA:添加金担子素A的缺尿嘧啶的SD培养基;SD/-Leu+AbA:添加金担子素A的缺亮氨酸SD培养基;SD/-Trp/-Leu:缺色氨酸和亮氨酸SD培养基;SD/-Trp/-Leu/-Ade/-Leu:缺色氨酸、组氨酸、腺嘌呤和亮氨酸SD培养基;SD/-Trp/-Leu/-Ade/-Leu/X-α-gal:添加X-α-Gal的缺色氨酸、组氨酸、腺嘌呤和亮氨酸SD培养基
Fig. 7 Yeast one-hybrid and yeast two-hybrid results a: CSS0013384 promoter self-activation detection. b: Results of yeast one-hybrid for CSS0013384. c: Results of yeast two-hybrid for CSS0036305. SD/-Leu+AbA: SD medium lacking leucine supplemented with aureobasidin A. SD/-Trp/-Leu: SD medium lacking tryptophan and leucine. SD/-Trp/-Leu/- Ade/-Leu: SD medium lacking tryptophan, histidine, adenine and leucine. SD/-Trp/-Leu/-Ade/-Leu/X-α-gal: SD medium lacking tryptophan, histidine, adenine and leucine supplemented with X-α-Gal
| [1] | 刘丁丁, 梅菊芬, 王君雅, 等. 茶树白化突变研究进展[J]. 中国茶叶, 2020, 42(4): 24-35. |
| Liu DD, Mei JF, Wang JY, et al. Research progress on albino trait of tea plant[J]. China Tea, 2020, 42(4): 24-35. | |
| [2] | 薄佳慧, 张杨玲, 宫连瑾, 等. 多组学视角下白化茶树次生代谢产物的研究进展[J/OL]. 分子植物育种, 2022. http://kns.cnki.net/kcms/detail/46.1068.S.20210412.1329.008.html. |
| Bo JH, Zhang YL, Gong LJ, et al. Research progress on secondary metabolites of albino tea from the perspective of multiomics[J/OL]. Mol Plant Breed, 2022. http://kns.cnki.net/kcms/detail/46.1068.S.20210412.1329.008.html. | |
| [3] | 范延艮, 赵秀秀, 王翰悦, 等. 黄金芽不同色泽叶片生理特性研究[J]. 茶叶科学, 2019, 39(5): 530-536. |
| Fan YG, Zhao XX, Wang HY, et al. Study on physiological characteristics of leaves with different colors of ‘Huangjinya’[J]. J Tea Sci, 2019, 39(5): 530-536. | |
| [4] | 吴全金. ‘白鸡冠’茶树响应光调控的基因差异及理化特征分析[D]. 福州: 福建农林大学, 2015. |
| Wu QJ. Gene differential analysis and physicochemical characteristics of Camellia sinensis cv. Baijiguan in response to light[D]. Fuzhou: Fujian Agriculture and Forestry University, 2015. | |
| [5] |
Li NN, Yang YP, Ye JH, et al. Effects of sunlight on gene expression and chemical composition of light-sensitive albino tea plant[J]. Plant Growth Regul, 2016, 78(2): 253-262.
doi: 10.1007/s10725-015-0090-6 URL |
| [6] | 周喆, 陈志丹, 吴全金, 等. 白鸡冠茶树CsPPH基因全长cDNA克隆与表达分析[J]. 茶叶科学, 2020, 40(1): 39-50. |
| Zhou Z, Chen ZD, Wu QJ, et al. Cloning and expression analysis of CsPPH gene in tea plant(Camellia sinensis)[J]. J Tea Sci, 2020, 40(1): 39-50. | |
| [7] | 王蔚, 郭雅玲. 白化茶品种的开发与应用[J]. 食品安全质量检测学报, 2017, 8(8): 3104-3110. |
| Wang W, Guo YL. Development and application of albino tea varieties[J]. J Food Saf & Qual, 2017, 8(8): 3104-3110. | |
| [8] | Wei K, Yu SP, Quan QL, et al. An integrative analysis of metabolomics, DNA methylation and RNA-Seq data reveals key genes involved in albino tea ‘Haishun 2’[J]. Beverage Plant Res, 2022, 2(1): 1-9. |
| [9] |
Li NN, Lu JL, Li QS, et al. Dissection of chemical composition and associated gene expression in the pigment-deficient tea cultivar ‘Xiaoxueya’ reveals an albino phenotype and metabolite formation[J]. Front Plant Sci, 2019, 10: 1543.
doi: 10.3389/fpls.2019.01543 URL |
| [10] | Du YY, Chen H, Zhong WL, et al. Effect of temperature on accumulation of chlorophylls and leaf ultrastructure of low temperature induced albino tea plant[J]. Afr J Biotechnol, 2008, 7(12): 1881-1885. |
| [11] |
Zhang QF, Tang DD, Liu MY, et al. Integrated analyses of the transcriptome and metabolome of the leaves of albino tea cultivars reveal coordinated regulation of the carbon and nitrogen metabolism[J]. Sci Hortic, 2018, 231: 272-281.
doi: 10.1016/j.scienta.2017.11.026 URL |
| [12] | Zhang QF, Liu MY, Ruan JY. Integrated transcriptome and metabolic analyses reveals novel insights into free amino acid metabolism in Huangjinya tea cultivar[J]. Front Plant Sci, 2017, 8: 291. |
| [13] | Wu QJ, Chen ZD, Sun WJ, et al. De novo sequencing of the leaf transcriptome reveals complex light-responsive regulatory networks in Camellia sinensis cv. Baijiguan[J]. Front Plant Sci, 2016, 7: 332. |
| [14] |
丁跃, 吴刚, 郭长奎. 植物叶绿素降解机制研究进展[J]. 生物技术通报, 2016, 32(11): 1-9.
doi: 10.13560/j.cnki.biotech.bull.1985.2016.11.001 |
|
Ding Y, Wu G, Guo CK. Research advance on chlorophyll degradation in plants[J]. Biotechnol Bull, 2016, 32(11): 1-9.
doi: 10.13560/j.cnki.biotech.bull.1985.2016.11.001 |
|
| [15] | 周丹, 罗灿, 于旭东, 等. 波罗蜜叶片突变体叶绿素含量测定和超微结构观察[J]. 热带作物学报, 2021, 42(10): 2935-2941. |
| Zhou D, Luo C, Yu XD, et al. Determination of chlorophyll content and observation of ultrastructure in leaves of mutants of Artocarpus heterophyllus[J]. Chin J Trop Crops, 2021, 42(10): 2935-2941. | |
| [16] |
陈伟, 吴昌荣, 鲁成, 等. 葡萄果实转色过程中线粒体呼吸代谢变化[J]. 核农学报, 2016, 30(6): 1227-1233.
doi: 10.11869/j.issn.100-8551.2016.06.1227 |
|
Chen W, Wu CR, Lu C, et al. Mitochondrial respiratory metabolism of grape during fruit coloring[J]. J Nucl Agric Sci, 2016, 30(6): 1227-1233.
doi: 10.11869/j.issn.100-8551.2016.06.1227 |
|
| [17] |
Lesnefsky EJ, Hoppel CL. Oxidative phosphorylation and aging[J]. Ageing Res Rev, 2006, 5(4): 402-433.
doi: 10.1016/j.arr.2006.04.001 pmid: 16831573 |
| [18] |
Rovira AG, Smith AG. PPR proteins - orchestrators of organelle RNA metabolism[J]. Physiol Plant, 2019, 166(1): 451-459.
doi: 10.1111/ppl.12950 pmid: 30809817 |
| [19] |
Okuda K, Chateigner-Boutin AL, Nakamura T, et al. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts[J]. Plant Cell, 2009, 21(1): 146-156.
doi: 10.1105/tpc.108.064667 URL |
| [20] |
Yagi Y, Tachikawa M, Noguchi H, et al. Pentatricopeptide repeat proteins involved in plant organellar RNA editing[J]. RNA Biol, 2013, 10(9): 1419-1425.
doi: 10.4161/rna.24908 pmid: 23669716 |
| [21] |
Zhang MY, Zhao YQ, Meng Y, et al. PPR proteins in the tea plant(Camellia sinensis)and their potential roles in the leaf color changes[J]. Sci Hortic, 2022, 293: 110745.
doi: 10.1016/j.scienta.2021.110745 URL |
| [22] | 刘丁丁, 王君雅, 汤榕津, 等. 茶树PPR基因家族全基因组鉴定及白化相关基因表达分析[J]. 茶叶科学, 2021, 41(2): 159-172. |
| Liu DD, Wang JY, Tang RJ, et al. Genome-wide identification of PPR gene family and expression analysis of albino related genes in tea plants[J]. J Tea Sci, 2021, 41(2): 159-172. | |
| [23] |
Du L, Zhang J, Qu SF, et al. The pentratricopeptide repeat protein pigment-defective mutant2 is involved in the regulation of chloroplast development and chloroplast gene expression in Arabidopsis[J]. Plant Cell Physiol, 2017, 58(4): 747-759.
doi: 10.1093/pcp/pcx004 URL |
| [24] |
Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines[J]. Cell, 1998, 92(3): 351-366.
doi: 10.1016/s0092-8674(00)80928-9 pmid: 9476895 |
| [25] |
Ditzel L, Löwe J, Stock D, et al. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT[J]. Cell, 1998, 93(1): 125-138.
pmid: 9546398 |
| [26] | Jackson-Constan D, Akita M, Keegstra K. Molecular chaperones involved in chloroplast protein import[J]. Biochim Biophys Acta, 2001, 1541(1/2): 102-113. |
| [27] |
Brocchieri L, Karlin S. Conservation among HSP60 sequences in relation to structure, function, and evolution[J]. Protein Sci, 2000, 9(3): 476-486.
pmid: 10752609 |
| [28] |
Jiang Q, Mei J, Gong XD, et al. Importance of the rice TCD9 encoding α subunit of chaperonin protein 60(Cpn60α)for the chloroplast development during the early leaf stage[J]. Plant Sci, 2014, 215/216: 172-179.
doi: 10.1016/j.plantsci.2013.11.003 URL |
| [29] |
Martel R, Cloney LP, Pelcher LE, et al. Unique composition of plastid chaperonin-60: alpha and beta polypeptide-encoding genes are highly divergent[J]. Gene, 1990, 94(2): 181-187.
pmid: 1979547 |
| [30] |
Peng LW, Shimizu H, Shikanai T. The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis[J]. J Biol Chem, 2008, 283(50): 34873-34879.
doi: 10.1074/jbc.M803207200 URL |
| [31] |
Hsu YW, Juan CT, Wang CM, et al. Mitochondrial heat shock protein 60s interact with what’s this factor 9 to regulate RNA splicing of ccmFC and rpl2[J]. Plant Cell Physiol, 2019, 60(1): 116-125.
doi: 10.1093/pcp/pcy199 URL |
| [32] |
Prasad TK, Stewart CR. cDNA clones encoding Arabidopsis thaliana and Zea mays mitochondrial chaperonin HSP60 and gene expression during seed germination and heat shock[J]. Plant Mol Biol, 1992, 18(5): 873-885.
pmid: 1349837 |
| [33] |
王涛, 王艺清, 周喆, 等. 白鸡冠茶树品种叶色白化相关共表达网络构建及潜在核心基因发掘[J/OL]. 应用与环境生物学报, 2022. DOI:10.19675/j.cnki.1006-687x.2022.11048.
doi: 10.19675/j.cnki.1006-687x.2022.11048 |
|
Wang T, Wang YQ, Zhou Z, et al. Mining genes related to leaf color change of Tea Plant(Camellia sinensis cv. Baijiguan)based on WGCNA[J/OL]. Chin J Appl Environ Biol, 2022. DOI:10.19675/j.cnki.1006-687x.2022.11048.
doi: 10.19675/j.cnki.1006-687x.2022.11048 |
|
| [34] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [35] | 张文婧, 林琳, 陈明杰, 等. 茶树CsAMT1s亚家族基因的克隆与表达[J]. 应用与环境生物学报, 2022, 28(1): 57-66. |
| Zhang WJ, Lin L, Chen MJ, et al. Molecular cloning and expression analysis of CsAMT1s gene subfamily in Camellia sinensis[J]. Chin J Appl Environ Biol, 2022, 28(1): 57-66. | |
| [36] |
Wang PJ, Zheng YC, Guo YC, et al. Widely targeted metabolomic and transcriptomic analyses of a novel albino tea mutant of “Rougui”[J]. Forests, 2020, 11(2): 229.
doi: 10.3390/f11020229 URL |
| [37] |
Xia EH, Tong W, Hou Y, et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation[J]. Mol Plant, 2020, 13(7): 1013-1026.
doi: S1674-2052(20)30134-9 pmid: 32353625 |
| [38] |
Yin P, Li QX, Yan CY, et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins[J]. Nature, 2013, 504(7478): 168-171.
doi: 10.1038/nature12651 |
| [39] |
Barkan A, Small I. Pentatricopeptide repeat proteins in plants[J]. Annu Rev Plant Biol, 2014, 65: 415-442.
doi: 10.1146/annurev-arplant-050213-040159 pmid: 24471833 |
| [40] | 王鑫伟, 安雅琪, 肖建伟. PPR蛋白调控质体基因表达和叶绿体发育的研究进展[J]. 植物生理学报, 2020, 56(12): 2510-2516. |
| Wang XW, An YQ, Xiao JW. Recent advances in PPR proteins regulation of plastid gene expression and chloroplast development[J]. Plant Physiol J, 2020, 56(12): 2510-2516. | |
| [41] | 王婉珍, 田发安, 任育军, 等. PPR蛋白在植物线粒体和叶绿体中功能的研究进展[J]. 福建农林大学学报: 自然科学版, 2018, 47(3): 257-266. |
| Wang WZ, Tian FA, Ren YJ, et al. Research progress on functions of PPR proteins in plant mitochondria and chloroplasts[J]. J Fujian Agric For Univ Nat Sci Ed, 2018, 47(3): 257-266. | |
| [42] | 陈龙. 水稻淡绿叶基因PGL12的克隆与功能分析[D]. 武汉: 华中农业大学, 2019. |
| Chen L. Cloning and functional analysis of PALE-GREEN LEAF 12 in rice[D]. Wuhan: Huazhong Agricultural University, 2019. | |
| [43] | 陈能刚. 水稻黄化转绿突变基因gry340的图位克隆与功能研究[D]. 雅安: 四川农业大学, 2018. |
| Chen NG. Map-based cloning and functional study of rice green-revertible yellow mutant gene gry340[D]. Ya'an: Sichuan Agricultural University, 2018. | |
| [44] | 暨国彪. 水稻白穗突变体基因WP3在水稻叶绿体与线粒体发育过程中的功能研究[D]. 北京: 中国科学院大学, 2011. |
| Ji GB. The function of rice white spike mutant gene WP3 in rice chloroplast and mitochondrial development[D]. Beijing: University of Chinese Academy of Sciences, 2011. | |
| [45] |
Leivar P, Monte E. PIFs: systems integrators in plant development[J]. Plant Cell, 2014, 26(1): 56-78.
doi: 10.1105/tpc.113.120857 URL |
| [46] |
Mizuno T, Oka H, Yoshimura F, et al. Insight into the mechanism of end-of-day far-red light(EODFR)-induced shade avoidance responses in Arabidopsis thaliana[J]. Biosci Biotechnol Biochem, 2015, 79(12): 1987-1994.
doi: 10.1080/09168451.2015.1065171 URL |
| [47] |
Zubo YO, Blakley IC, Franco-Zorrilla JM, et al. Coordination of chloroplast development through the action of the GNC and GLK transcription factor families[J]. Plant Physiol, 2018, 178(1): 130-147.
doi: 10.1104/pp.18.00414 pmid: 30002259 |
| [48] |
Vitlin Gruber A, Vugman M, Azem A, et al. Reconstitution of pure chaperonin hetero-oligomer preparations in vitro by temperature modulation[J]. Front Mol Biosci, 2018, 5: 5.
doi: 10.3389/fmolb.2018.00005 pmid: 29435453 |
| [1] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [2] | 赵光绪, 杨合同, 邵晓波, 崔志豪, 刘红光, 张杰. 一株高效溶磷产红青霉培养条件优化及其溶磷特性[J]. 生物技术通报, 2023, 39(9): 71-83. |
| [3] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [4] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [5] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [6] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [7] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [8] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [9] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [10] | 庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115. |
| [11] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [12] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [13] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [14] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [15] | 尤垂淮, 谢津津, 张婷, 崔天真, 孙欣路, 臧守建, 武奕凝, 孙梦瑶, 阙友雄, 苏亚春. 钩吻脂氧合酶基因 GeLOX1 的鉴定及低温胁迫表达分析[J]. 生物技术通报, 2023, 39(11): 318-327. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||