








生物技术通报 ›› 2023, Vol. 39 ›› Issue (6): 316-324.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1255
收稿日期:2022-10-11
出版日期:2023-06-26
发布日期:2023-07-07
通讯作者:
董聪同时为本文通讯作者作者简介:董聪,女,硕士,助理研究员,研究方向:酶制剂;E-mail: dongcong24@126.com;
基金资助:
DONG Cong( ), GAO Qing-hua, WANG Yue, LUO Tong-yang, WANG Qing-qing
), GAO Qing-hua, WANG Yue, LUO Tong-yang, WANG Qing-qing
Received:2022-10-11
Published:2023-06-26
Online:2023-07-07
摘要:
为了提高了FAD依赖的葡萄糖脱氢酶在毕赤酵母中的表达量,本研究通过G418浓度梯度筛选,构建含有不同分子伴侣基因拷贝数共表达毕赤酵母重组菌株实现高效表达,优化摇瓶发酵条件。经G418浓度梯度筛选,得到1株高产重组菌,在此基础上分别构建了HAC1、Ero1、PDI三种分子伴侣1-4拷贝共表达重组菌株,RT-qPCR检测结果符合预期。试管水平酶活表明3拷贝PDI、3拷贝HAC1和2拷贝Ero1分别比1拷贝提高83%、77%和51%。总体比较而言,共表达3拷贝PDI的重组菌株酶活最高,比对照提高198%,优化摇瓶发酵条件表明培养基初始pH 7.0,摇瓶装液量50 mL,甲醇添加量1%,诱导温度28℃时,效果最佳。通过抗生素浓度筛选、增加分子伴侣的基因拷贝数策略可以提高FAD-GDH在毕赤酵母中的发酵产量。
董聪, 高庆华, 王玥, 罗同阳, 王庆庆. 基于联合策略提高FAD依赖的葡萄糖脱氢酶的酵母表达[J]. 生物技术通报, 2023, 39(6): 316-324.
DONG Cong, GAO Qing-hua, WANG Yue, LUO Tong-yang, WANG Qing-qing. Increasing the Expression of FAD-dependent Glucose Dehydrogenase by Recombinant Pichia pastoris Using a Combined Strategy[J]. Biotechnology Bulletin, 2023, 39(6): 316-324.
| 引物名称 Primer name | 基因 Gene | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| H- F | HAC1 | CTCGCAGATCCTTGACTGAGGATCTGGACGAAG |
| H- R | CCTCAGTCAAGGATCTGCGAGTGGATGTAGATG | |
| E- F | Ero1 | TGGAGAAACAAATCTTTTACCGATTGGTTTCTG |
| E- R | GGTAAAAGATTTGTTTCTCCAAACAGAGGTCTTC | |
| P1-F | PDI | CTGCTGCCGAAATCTTAAAGGACAATGAGCAGG |
| P1-R | CCTTTAAGATTTCGGCAGCAGAAACAAGTTCAG | |
| P2-F | TTGACGGATCTACCTTCAAATCATATGCTGAAG | |
| P2-F | ATTTGAAGGTAGATCCGTCAATGTCTCCAAATAG |
表1 突变引物序列
Table 1 Mutation primer sequences
| 引物名称 Primer name | 基因 Gene | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| H- F | HAC1 | CTCGCAGATCCTTGACTGAGGATCTGGACGAAG |
| H- R | CCTCAGTCAAGGATCTGCGAGTGGATGTAGATG | |
| E- F | Ero1 | TGGAGAAACAAATCTTTTACCGATTGGTTTCTG |
| E- R | GGTAAAAGATTTGTTTCTCCAAACAGAGGTCTTC | |
| P1-F | PDI | CTGCTGCCGAAATCTTAAAGGACAATGAGCAGG |
| P1-R | CCTTTAAGATTTCGGCAGCAGAAACAAGTTCAG | |
| P2-F | TTGACGGATCTACCTTCAAATCATATGCTGAAG | |
| P2-F | ATTTGAAGGTAGATCCGTCAATGTCTCCAAATAG |
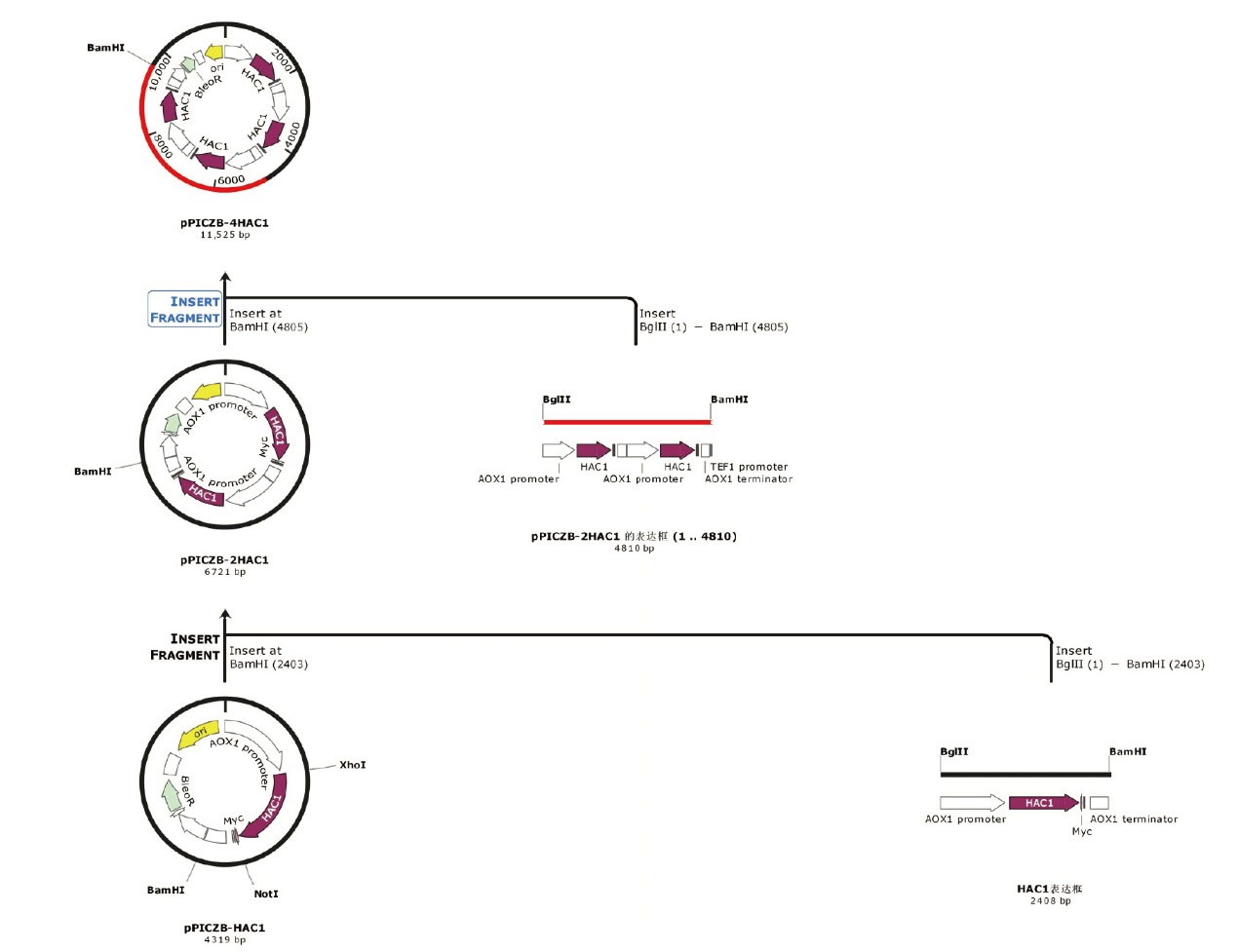
图1 分子伴侣HAC1多拷贝表达盒载体的构建策略图
Fig. 1 Schematic representation of the recombinant vectors harboring multi-copy expression cassette for molecular chaperones HAC1
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| RT GAP up | CCAGCGGCAACAAGATCAAC |
| RT GAP down | CTCCTCGTTGACACCGACAA |
| RT GDH up | GGCAATACCAATCTGTCAACCAACCTTA |
| RT GDH down | AACCAGTGTTACCGATAGTTTCCCAAGC |
| RT HAC1 up | CTGAATATGACGACGAAGAA |
| RT HAC1 down | CTCCTGCTTGATAGATGTG |
| RT PDI up | ACCACATTTTACGGAGTTGCCGGT |
| RT PDI down | CCTCGCCAGGTCTGACAAGCA |
| RT Ero1 up | ATGAGGATAGTAAGGAGCGTAGC |
| RT Ero1 down | GTTGACCAGTTCCACCAGTTTG |
表2 RT-qPCR所用引物
Table 2 Primers used in RT-qPCR
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| RT GAP up | CCAGCGGCAACAAGATCAAC |
| RT GAP down | CTCCTCGTTGACACCGACAA |
| RT GDH up | GGCAATACCAATCTGTCAACCAACCTTA |
| RT GDH down | AACCAGTGTTACCGATAGTTTCCCAAGC |
| RT HAC1 up | CTGAATATGACGACGAAGAA |
| RT HAC1 down | CTCCTGCTTGATAGATGTG |
| RT PDI up | ACCACATTTTACGGAGTTGCCGGT |
| RT PDI down | CCTCGCCAGGTCTGACAAGCA |
| RT Ero1 up | ATGAGGATAGTAAGGAGCGTAGC |
| RT Ero1 down | GTTGACCAGTTCCACCAGTTTG |
| 菌株 Strain | GDH基因拷贝数 Copy number of GDH | HAC1基因拷贝数 Copy number of HAC1 | PDI基因拷贝数 Copy number of PDI | Ero1基因拷贝数 Copy number of Ero1 |
|---|---|---|---|---|
| CK(X33/GDH) | 3.20±0.20 | Endogenous 1 | Endogenous 1 | Endogenous 1 |
| CK-H | 3.20±0.20 | 1.10±0.09 | Endogenous 1 | Endogenous 1 |
| CK-2H | 3.20±0.20 | 2.08±0.26 | Endogenous 1 | Endogenous 1 |
| CK-3H | 3.20±0.20 | 2.95±0.11 | Endogenous 1 | Endogenous 1 |
| CK-4H | 3.20±0.20 | 4.35±0.21 | Endogenous 1 | Endogenous 1 |
| CK-P | 3.20±0.20 | Endogenous 1 | 1.02±0.11 | Endogenous 1 |
| CK-2P | 3.20±0.20 | Endogenous 1 | 2.32±0.19 | Endogenous 1 |
| CK-3P(X33/GDH-3PDI) | 3.20±0.20 | Endogenous 1 | 3.006±0.20 | Endogenous 1 |
| CK-4P | 3.20±0.20 | Endogenous 1 | 4.42±0.29 | Endogenous 1 |
| CK-E | 3.20±0.20 | Endogenous 1 | Endogenous 1 | 1.21±0.13 |
| CK-2E | 3.20±0.20 | Endogenous 1 | Endogenous 1 | 1.98±0.34 |
| CK-3E | 3.20±0.20 | Endogenous 1 | Endogenous 1 | 3.22±0.17 |
| CK-4E | 3.20±0.20 | Endogenous 1 | Endogenous 1 | 4.19±0.20 |
表3 RT-qPCR检测基因拷贝数
Table 3 Copy numbers of genes detected by RT-qPCR
| 菌株 Strain | GDH基因拷贝数 Copy number of GDH | HAC1基因拷贝数 Copy number of HAC1 | PDI基因拷贝数 Copy number of PDI | Ero1基因拷贝数 Copy number of Ero1 |
|---|---|---|---|---|
| CK(X33/GDH) | 3.20±0.20 | Endogenous 1 | Endogenous 1 | Endogenous 1 |
| CK-H | 3.20±0.20 | 1.10±0.09 | Endogenous 1 | Endogenous 1 |
| CK-2H | 3.20±0.20 | 2.08±0.26 | Endogenous 1 | Endogenous 1 |
| CK-3H | 3.20±0.20 | 2.95±0.11 | Endogenous 1 | Endogenous 1 |
| CK-4H | 3.20±0.20 | 4.35±0.21 | Endogenous 1 | Endogenous 1 |
| CK-P | 3.20±0.20 | Endogenous 1 | 1.02±0.11 | Endogenous 1 |
| CK-2P | 3.20±0.20 | Endogenous 1 | 2.32±0.19 | Endogenous 1 |
| CK-3P(X33/GDH-3PDI) | 3.20±0.20 | Endogenous 1 | 3.006±0.20 | Endogenous 1 |
| CK-4P | 3.20±0.20 | Endogenous 1 | 4.42±0.29 | Endogenous 1 |
| CK-E | 3.20±0.20 | Endogenous 1 | Endogenous 1 | 1.21±0.13 |
| CK-2E | 3.20±0.20 | Endogenous 1 | Endogenous 1 | 1.98±0.34 |
| CK-3E | 3.20±0.20 | Endogenous 1 | Endogenous 1 | 3.22±0.17 |
| CK-4E | 3.20±0.20 | Endogenous 1 | Endogenous 1 | 4.19±0.20 |
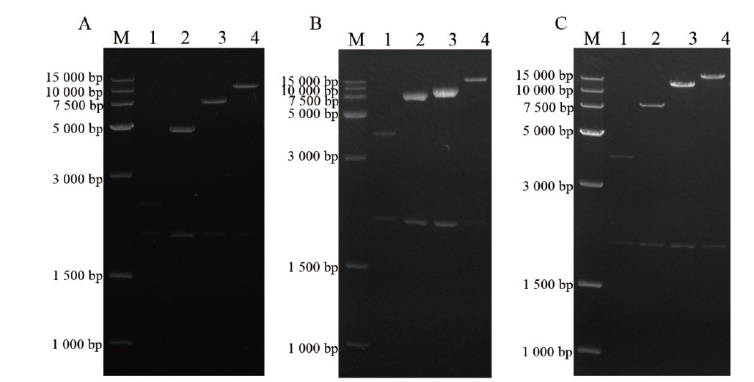
图2 pPICZB-HAC1n(n=1,2,3或4)(A)、pPICZB-Ero1n(n=1,2,3或4)(B)、pPICZB-PDIn(n=1,2,3或4)(C)经双酶切后的琼脂糖凝胶电泳分析 M:DNA分子量标准;1-4:经Bgl II 和 BamH I双酶切后的pPICZB-HAC1/Ero1/PDIn(n=1,2,3或4)
Fig. 2 Agarose gel electrophoresis analysis for pPICZB-HAC1n(n=1,2,3或4)(A),pPICZB-Ero1n(n=1,2,3或4)(B),pPICZB-PDIn(n=1,2,3或4)(C)digested with double enzymes M: DNA ladder; lanes 1-4: pPICZB-HAC1/Ero1/PDIn(n=1,2,3或4)digested with BamH I and Bgl II
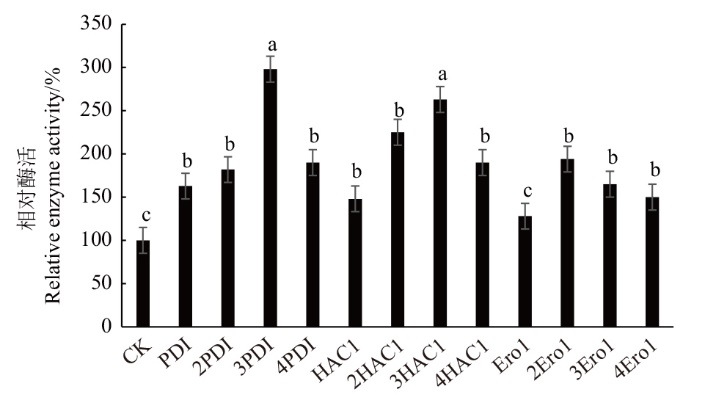
图3 试管水平不同分子伴侣共表达重组菌株相对酶活 图中误差线表示标准偏差,小写字母表示不同菌株酶活差异达到(P<0.05)显著水平,下同
Fig. 3 Relative enzyme activities of recombinant strains co-expressed with different molecular chaperones at test tube level The error line in the figure refers to the standard deviation. The lowercase letters indicate that the difference among different strains reached a significant level(P<0.05). The same below
| [1] |
Fapyane D, Lee SJ, Kang SH, et al. High performance enzyme fuel cells using a genetically expressed FAD-dependent glucose dehydrogenase α-subunit of Burkholderia cepacia immobilized in a carbon nanotube electrode for low glucose conditions[J]. Phys Chem Chem Phys, 2013, 15(24): 9508-9512.
doi: 10.1039/c3cp51864g pmid: 23695009 |
| [2] |
Southcott M, MacVittie K, Halámek J, et al. A pacemaker powered by an implantable biofuel cell operating under conditions mimicking the human blood circulatory system—battery not included[J]. Phys Chem Chem Phys, 2013, 15(17): 6278-6283.
doi: 10.1039/c3cp50929j pmid: 23519144 |
| [3] |
Tang Z, Louie RF, Lee JH, et al. Oxygen effects on glucose meter measurements with glucose dehydrogenase- and oxidase-based test strips for point-of-care testing[J]. Crit Care Med, 2001, 29(5): 1062-1070.
pmid: 11378622 |
| [4] |
Iwasa H, Ozawa K, Sasaki N, et al. Fungal FAD-dependent glucose dehydrogenases concerning high activity, affinity, and thermostability for maltose-insensitive blood glucose sensor[J]. Biochem Eng J, 2018, 140: 115-122.
doi: 10.1016/j.bej.2018.09.014 URL |
| [5] |
Yang YF, Huang L, Wang JF, et al. Efficient expression, purification, and characterization of a novel FAD-dependent glucose dehydrogenase from Aspergillus terreus in Pichia pastoris[J]. J Microbiol Biotechnol, 2014, 24(11): 1516-1524.
doi: 10.4014/jmb.1401.01061 URL |
| [6] |
Yang YF, Huang L, Wang JF, et al. Expression, characterization and mutagenesis of an FAD-dependent glucose dehydrogenase from Aspergillus terreus[J]. Enzyme Microb Technol, 2015, 68: 43-49.
doi: 10.1016/j.enzmictec.2014.10.002 URL |
| [7] |
Hohenblum H, Gasser B, Maurer M, et al. Effects of gene dosage, promoters, and substrates on unfolded protein stress of recombinant Pichia pastoris[J]. Biotechnol Bioeng, 2004, 85(4): 367-375.
doi: 10.1002/bit.10904 pmid: 14755554 |
| [8] |
Aw R, Polizzi KM. Can too many copies spoil the broth?[J]. Microb Cell Fact, 2013, 12: 128.
doi: 10.1186/1475-2859-12-128 pmid: 24354594 |
| [9] | 黄猛猛. 利用组合策略促进毕赤酵母异源表达α-淀粉酶[D]. 上海: 华东理工大学, 2016. |
| Huang MM. Enhancing expression of α-amylase in Pichia pastoris by combined strategies[D]. Shanghai: East China University of Science and Technology, 2016. | |
| [10] |
Sun J, Jiang J, Zhai XY, et al. Coexpression of Kex2 endoproteinase and Hac1 transcription factor to improve the secretory expression of bovine lactoferrin in Pichia pastoris[J]. Biotechnol Bioprocess Eng, 2019, 24(6): 934-941.
doi: 10.1007/s12257-019-0176-5 |
| [11] |
Yu P, Zhu Q, Chen KF, et al. Improving the secretory production of the heterologous protein in Pichia pastoris by focusing on protein folding[J]. Appl Biochem Biotechnol, 2015, 175(1): 535-548.
doi: 10.1007/s12010-014-1292-5 URL |
| [12] |
Qiang L, Wang H, Farmer SR. Adiponectin secretion is regulated by SIRT1 and the endoplasmic Reticulum oxidoreductase Ero1-L alpha[J]. Mol Cell Biol, 2007, 27(13): 4698-4707.
pmid: 17452443 |
| [13] |
Shen Q, Wu M, Wang HB, et al. The effect of gene copy number and co-expression of chaperone on production of albumin fusion proteins in Pichia pastoris[J]. Appl Microbiol Biotechnol, 2012, 96(3): 763-772.
doi: 10.1007/s00253-012-4337-0 pmid: 22885695 |
| [14] | 闵琪. 耐热甘露聚糖酶ManA在毕赤酵母中的高效表达[D]. 武汉: 中南民族大学, 2020. |
| Min Q. High-lever expression of thermostable mannanase ManA in Pichia pastoris[D]. Wuhan: South-central University for Nationalities, 2020. | |
| [15] | 钱晓芬, 吴涛, 赵理想, 等. 基因拷贝数对重组毕赤酵母的牛乳铁蛋白功能片段表达及细胞存活率的影响[J]. 食品与发酵工业, 2021, 47(4): 1-6. |
| Qian XF, Wu T, Zhao LX, et al. Effect of gene copy number on the expression of bovine lactoferrin functional fragment and cell survival in recombinant Pichia pastoris[J]. Food Ferment Ind, 2021, 47(4): 1-6. | |
| [16] |
Huang MM, Gao YY, Zhou XS, et al. Regulating unfolded protein response activator HAC1p for production of thermostable raw-starch hydrolyzing α-amylase in Pichia pastoris[J]. Bioprocess Biosyst Eng, 2017, 40(3): 341-350.
doi: 10.1007/s00449-016-1701-y URL |
| [17] |
Inan M, Aryasomayajula D, Sinha J, et al. Enhancement of protein secretion in Pichia pastoris by overexpression of protein disulfide isomerase[J]. Biotechnol Bioeng, 2006, 93(4): 771-778.
doi: 10.1002/(ISSN)1097-0290 URL |
| [18] | 祁丽, 姜宁, 张爱忠, 等. 抗菌肽多基因表达技术与策略[J]. 黑龙江畜牧兽医, 2016(15): 52-56. |
| Qi L, Jiang N, Zhang AZ, et al. Multi-gene expression technology and strategy of antibacterial peptides[J]. Heilongjiang Animal Sci Vet Med, 2016(15): 52-56. | |
| [19] |
董聪, 高庆华, 王玥, 等. 基于密码子优化的FAD依赖葡萄糖脱氢酶在毕赤酵母中的高效表达及酶学性质[J]. 生物技术通报, 2019, 35(7): 114-120.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-0941 |
| Dong C, Gao QH, Wang Y, et al. Expression and enzymatic characterization of Codon-optimized FAD-dependent glucose dehydrogenase in Pichia pastoris[J]. Biotechnol Bull, 2019, 35(7): 114-120. | |
| [20] |
Abad S, Kitz K, Hörmann A, et al. Real-time PCR-based determination of gene copy numbers in Pichia pastoris[J]. Biotechnol J, 2010, 5(4): 413-420.
doi: 10.1002/biot.v5:4 URL |
| [21] |
Li D, Wu JC, Chen J, et al. Optimized expression of classical swine fever virus E2 protein via combined strategy in Pichia pastoris[J]. Protein Expr Purif, 2020, 167: 105527.
doi: 10.1016/j.pep.2019.105527 URL |
| [22] |
Sygmund C, Staudigl P, Klausberger M, et al. Heterologous overexpression of Glomerella cingulata FAD-dependent glucose dehydrogenase in Escherichia coli and Pichia pastoris[J]. Microb Cell Fact, 2011, 10: 106.
doi: 10.1186/1475-2859-10-106 |
| [23] | 马银鹏, 王玉文, 党阿丽, 等. 毕赤酵母表达系统研究进展[J]. 黑龙江科学, 2013, 4(9): 27-31. |
| Ma YP, Wang YW, Dang AL, et al. Advances of Pichia pastoris expression system[J]. Heilongjiang Sci, 2013, 4(9): 27-31. | |
| [24] |
Hwang J, Qi L. Quality control in the endoplasmic Reticulum: crosstalk between ERAD and UPR pathways[J]. Trends Biochem Sci, 2018, 43(8): 593-605.
doi: 10.1016/j.tibs.2018.06.005 URL |
| [25] |
Yang J, Lu ZP, Chen JW, et al. Effect of cooperation of chaperones and gene dosage on the expression of porcine PGLYRP-1 in Pichia pastoris[J]. Appl Microbiol Biotechnol, 2016, 100(12): 5453-5465.
doi: 10.1007/s00253-016-7372-4 pmid: 26883349 |
| [26] |
Kim JM, Jang SA, Yu BJ, et al. High-level expression of an antimicrobial peptide histonin as a natural form by multimerization and furin-mediated cleavage[J]. Appl Microbiol Biotechnol, 2008, 78(1): 123-130.
pmid: 18094965 |
| [27] |
Cos O, Serrano A, Montesinos JL, et al. Combined effect of the methanol utilization(Mut)phenotype and gene dosage on recombinant protein production in Pichia pastoris fed-batch cultures[J]. J Biotechnol, 2005, 116(4): 321-335.
doi: 10.1016/j.jbiotec.2004.12.010 URL |
| [28] |
Song XP, Shao CS, Guo YG, et al. Improved the expression level of active transglutaminase by directional increasing copy of mtg gene in Pichia pastoris[J]. BMC Biotechnol, 2019, 19(1): 54.
doi: 10.1186/s12896-019-0542-6 |
| [29] | 焦梁成. 毕赤酵母分子操作方法改进及其在米根霉脂肪酶高效表达中的应用[D]. 武汉: 华中科技大学, 2019. |
| Jiao LC. Improvement of molecular manipulation method of Pichia pastoris and its application in high-level expression of Rhizopus oryzae lipase[D]. Wuhan: Huazhong University of Science and Technology, 2019. |
| [1] | 赵思佳, 王晓璐, 孙纪录, 田健, 张杰. 代谢工程改造毕赤酵母生产赤藓糖醇[J]. 生物技术通报, 2023, 39(8): 137-147. |
| [2] | 赵昕, 杜玉瑶, 殷子扬, 毛淑红. 胆固醇7α-羟化酶在毕赤酵母中的异源表达[J]. 生物技术通报, 2023, 39(10): 304-310. |
| [3] | 王玥, 高庆华, 董聪, 罗同阳, 王庆庆. 密码子优化的吡喃糖氧化酶基因在毕赤酵母中的表达[J]. 生物技术通报, 2022, 38(4): 269-277. |
| [4] | 杨威, 伍茜, 程建国, 罗燕, 王印, 杨泽晓, 姚学萍. 林麝干扰素α基因克隆、表达及转录调控分析[J]. 生物技术通报, 2022, 38(1): 194-204. |
| [5] | 廖兆民, 蔡俊, 林建国, 杜馨, 王常高. 黑曲霉葡萄糖氧化酶基因在毕赤酵母中的表达及产酶条件的优化[J]. 生物技术通报, 2021, 37(6): 97-107. |
| [6] | 杨悦, 陶妍, 谢晶, 钱韻芳. 基于重组毕赤酵母的草鱼C型溶菌酶生物合成及其抑菌活性[J]. 生物技术通报, 2021, 37(12): 169-179. |
| [7] | 赵震, 王莎莎, 吕星星, 陶妍, 谢晶, 钱韻芳. 重组毕赤酵母产青蛤Mytimacin抗菌肽的表达研究[J]. 生物技术通报, 2020, 36(5): 150-158. |
| [8] | 闵琪, 高子涵, 姚银, 张华山, 熊海容, 张莉. 共表达HAC1和分子伴侣基因对甘露聚糖酶在毕赤酵母中表达的影响[J]. 生物技术通报, 2020, 36(5): 159-168. |
| [9] | 李雅楠, 余利红, 陈新美, 杨浩萌, 黄火清. 来源于Penicillium sp.C1的水产用中性植酸酶基因在毕赤酵母中的表达及性质研究[J]. 生物技术通报, 2020, 36(2): 134-141. |
| [10] | 张亚莉, 陶妍, 谢晶, 钱韻芳. 厚壳贻贝Mytilin-1成熟肽在毕赤酵母中的重组表达及其抑菌活性[J]. 生物技术通报, 2019, 35(7): 54-60. |
| [11] | 侯成林, 杨艳坤, 陈嘉荔, 白仲虎. Mxr1磷酸化水平受Ptp调控机理的初步研究[J]. 生物技术通报, 2019, 35(7): 108-113. |
| [12] | 董聪, 高庆华, 王玥, 罗同阳. 基于密码子优化的FAD依赖葡萄糖脱氢酶在毕赤酵母中的高效表达及酶学性质[J]. 生物技术通报, 2019, 35(7): 114-120. |
| [13] | 刘进兰, 杨雪, 李双双, 张玉明, 柳峰松, 唐婷, 李红权. 家蝇抗菌肽Domesticin在毕赤酵母中的表达及抑菌活性检测[J]. 生物技术通报, 2019, 35(2): 109-115. |
| [14] | 刘思阳, 王岩, 付玉芹, 胡祥坤, 王茗宇, 卢天成, 王秀然. 毕赤酵母糖基化对布鲁菌P39抗原诱导巨噬细胞吞噬的影响[J]. 生物技术通报, 2018, 34(9): 237-243. |
| [15] | 高庆华, 董聪, 王玥, 胡美荣, 王庆庆, 王云鹏, 罗同阳, 刘蕾. 共表达分子伴侣PDI和Ero1对葡萄糖氧化酶在毕赤酵母中表达的影响[J]. 生物技术通报, 2018, 34(7): 174-179. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||