








生物技术通报 ›› 2023, Vol. 39 ›› Issue (7): 1-12.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0051
• 综述与专论 • 下一篇
陈晓1( ), 于茗兰1, 吴隆坤2, 郑晓明3,4,5, 逄洪波1(
), 于茗兰1, 吴隆坤2, 郑晓明3,4,5, 逄洪波1( )
)
收稿日期:2023-01-19
出版日期:2023-07-26
发布日期:2023-08-17
通讯作者:
逄洪波,女,博士,副教授,研究方向 :植物逆境分子生物学;E-mail: panghb@synu.edu.cn作者简介:陈晓,女,硕士研究生,研究方向:生物化学与分子生物学;E-mail: 19861602091@163.com
基金资助:
CHEN Xiao1( ), YU Ming-lan1, WU Long-kun2, ZHENG Xiao-ming3,4,5, PANG Hong-bo1(
), YU Ming-lan1, WU Long-kun2, ZHENG Xiao-ming3,4,5, PANG Hong-bo1( )
)
Received:2023-01-19
Published:2023-07-26
Online:2023-08-17
摘要:
低温限制植物生长区域,决定种植范围;导致作物产量和品质下降,严重可能造成植株死亡。为了抵抗低温胁迫,植物进化出了复杂的防御机制。lncRNA是存在于细胞核和细胞质中、由体内基因组转录产生的一类转录本。近年来,已证实lncRNA可以通过多聚腺苷酸化、与某些蛋白酶协同作用、与miRNA竞争性结合等方式来响应植物低温胁迫。本文就lncRNA的定义、来源、分类以及低温条件下lncRNA在模式植物拟南芥和农经作物中的研究进展进行了总结,期望为植物耐低温机理研究及植物耐冷分子育种提供参考。
陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12.
CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant[J]. Biotechnology Bulletin, 2023, 39(7): 1-12.

图1 lncRNA的结构及来源 A:lncRNA的基因结构;B:lncRNA的形成机制;(1)阅读框插入到蛋白质编码基因的内含子中,插入的序列与之前的序列重新整合形成lncRNA;(2)染色体重新组合,2个远距离(>10 Mb)的蛋白质编码基因的非编码区串联在一起,形成含有多个外显子的lncRNA;(3)非编码基因的反转录转座子以RNA为媒介,通过反转录,以复制粘贴的方式在基因组的新位置产生一个新的拷贝,与之前的非编码序列一起形成lncRNA;(4)连续重复事件在非编码RNA内形成相邻的重复序列从而产生新的lncRNA;(5)转位因子插入到非编码基因中,与之前的序列一起形成lncRNA
Fig. 1 Structure and origin of lncRNA A: Gene structure of lncRNA. B: The mechanism of lncRNA formation.(1)The reading frame inserted into the intron of the protein-coding gene, and the inserted sequence is reintegrated with the previous sequence to form lncRNA.(2)Chromosome recombination, two long-distance(>10 Mb)non-coding regions of protein-coding genes connected together to form lncRNA containing multiple exons.(3)Non-coding gene retrotransposons use RNA as a medium to create a new copy at a new location in the genome through reverse transcription and paste it by copying and pasting, forming a lncRNA in conjunction with the previous non-coding sequences.(4)Tandem repeat events generate adjacent repeat sequences within non-coding RNA to produce new lncRNA.(5)Transposable elements inserted into non-coding genes, which forms a lncRNA in conjunction with the previous sequence
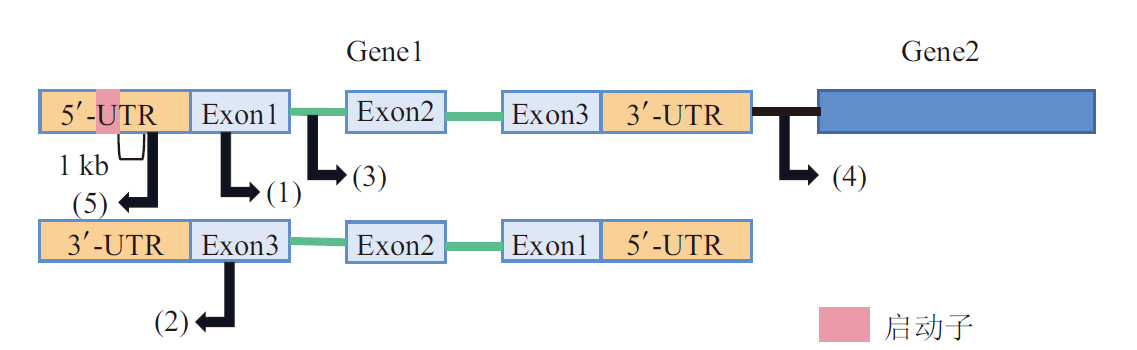
图2 基于基因组位置的lncRNA分类 (1)正义lncRNA,从蛋白质编码基因的正义链转录形成;(2)反义lncRNA,从蛋白质编码基因的反义链向反方向转录形成;(3)内含子lncRNA,从蛋白质编码基因的内含子处转录形成;(4)基因间lncRNA,由两个基因之间的区域转录形成;(5)双向lncRNA,从蛋白质编码基因的启动子附近向与蛋白质编码基因的反方向转录形成
Fig. 2 Classification of lncRNAs based on genomic locations (1)Sense lncRNA is transcribed from the sense strand of protein-coding genes.(2)Antisense lncRNA is transcribed from the antisense strand of protein-coding genes in the opposite direction.(3)Intron lncRNA is transcribed from the intronic region of protein-coding genes.(4)Intergenic lncRNA is transcribed from the region between two genes.(5)Bidirectional lncRNA is transcribed in the opposite direction of protein-coding genes from the promoter region.
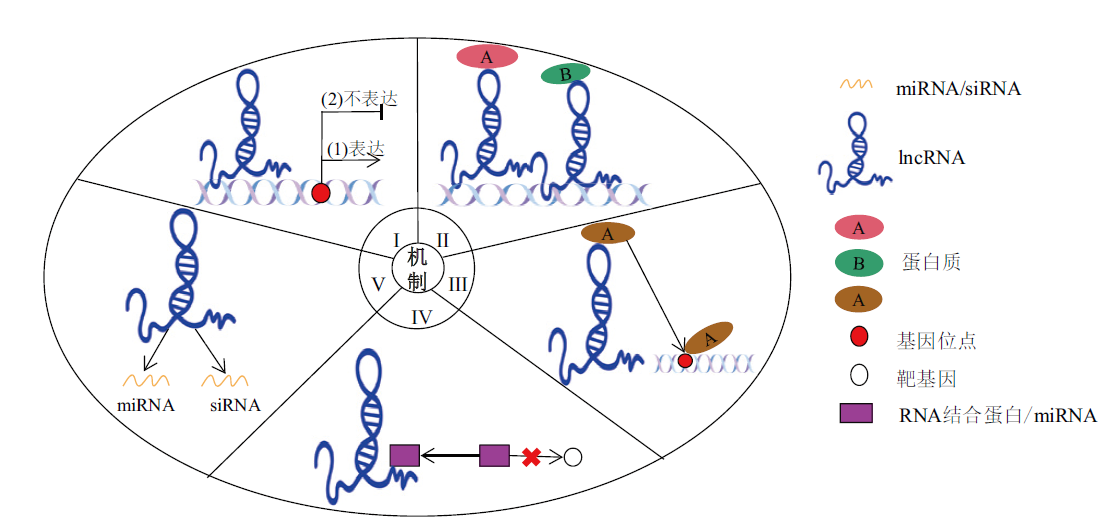
图3 lncRNA的作用机制示意图 I:信号分子模型,lncRNA作为信号分子控制邻近基因,决定其是否表达;II:支架分子模型,lncRNA作为支架结合多种不同蛋白质发挥作用;III:引导分子模型,lncRNA与蛋白质结合并将其引导到指定位置发挥作用;IV:诱饵分子模型,lncRNA诱导RNA结合蛋白或miRNA与其结合,使它们远离靶基因;V:lncRNA作为miRNAs和siRNAs生物合成的前体
Fig. 3 Schematic diagram of lncRNA action mechanisms I: Signal molecule model, as signaling molecules, lncRNAs control neighboring genes to determine whether they are expressed; II: scaffold molecular model, the lncRNAs act as scaffolding molecules for a variety of proteins; III: guided molecular model, proteins are directed to specific sites for action by lncRNAs; IV: the bait molecule model, induced RNA-binding proteins or miRNAs bind to lncRNA, preventing them from binding to target genes; V: lncRNA acts as a precursor for the biosynthesis of miRNAs and siRNAs
| lncRNA的名称 Name of lncRNA | 物种 Species | 功能 Function | 机制 Mechanism | 参考文献 Reference |
|---|---|---|---|---|
| COLDAIR | 拟南芥 A. thaliana | 参与春化作用 Involved in vernalization | 信号、引导分子,抑制FLC的表达 Signal, guide molecules, and inhibit the expression of FLC | [ |
| COLDWRAP | 拟南芥 A. thaliana | 参与春化作用 Involved in vernalization | 抑制FLC的表达 Inhibit the expression of FLC | [ |
| COOLAIR | 拟南芥 A. thaliana | 参与春化作用 Involved in vernalization | 增加H3K27me3和降低H3K36me3转录来抑制FLC Inhibition of FLC by increasing H3K27me3 and decreasing H3K36me3 transcription | [ |
| Svalka | 拟南芥 A. thaliana | 参与冷胁迫 Involved in cold stress | Svalka-asCBF1级联控制CBF1表达 Svalka-asCBF1 cascade controls CBF1 expression | [ |
| CIL1 | 拟南芥 A. thaliana | 参与冷胁迫 Involved in cold stress | 影响活性氧或渗透调节物质来响应冷胁迫 Responding to cold stress by affecting reactive oxygen species or osmotic regulators | [ |
| APOLO | 拟南芥 A. thaliana | 寒冷条件调节根毛伸长 Regulates root hair elongation under cold conditions | 与转录因子WRKY42相互作用 Interacting with the TF WRKY42 | [ |
| TE-lincRNA11195 | 拟南芥 A. thaliana | 参与冷胁迫 Involved in cold stress | 水杨酸刺激反应基因可能是潜在靶标 Salicylic acid-induced response genes may be potential targets | [ |
| DPA lncRNAs | 水稻 O. sativa | 参与冷胁迫 Involved in cold stress | 多聚腺苷酸化来响应冷胁迫 Polyadenylation in response to cold stress | [ |
| lncRNA-Chr03G0008 | 水稻 O. sativa | 参与冷胁迫 Involved in cold stress | 在幼苗中表达响应冷胁迫 Expressing response to cold stress in seedlings | [ |
| lncRNA-SVR | 水稻 O. sativa | 低温下与种子活力相关 Associated with seed viability under low temperature | 顺式基因SAUR家族成员相互作用 Interacting with the cis-gene family member SAUR | [ |
| lncR9A, lncR117, lncR616 | 东农冬麦1号 T. aestivum(Dn1) | 参与冷胁迫 Involved in cold stress | 竞争性结合miR398间接调节CSD1的表达 Indirectly regulates the expression of CSD1 by competitively binding miR398 | [ |
| Traes_2BS_7A04BF5D5 | 小麦 Durum wheat | 参与冷胁迫 Involved in cold stress | 以WCOR413冷驯化基因为靶标响应冷胁迫 Responding to cold stress targeting the cold acclimation gene WCOR413 | [ |
| CRR5 | 木薯 M. esculenta | 参与冷胁迫 Involved in cold stress | 与蛋白激酶基因协同表达响应低温胁迫 Co-expression with protein kinase gene in response to low temperature stress | [ |
| CRIR1 | 木薯 M. esculenta | 参与冷胁迫 Involved in cold stress | 招募MeCSP5来提高mRNA的翻译效率 Recruit MeCSP5 to improve the translation efficiency of mRNA | [ |
| XH123 | 棉花 G. hirsutum | 参与冷胁迫 Involved in cold stress | 基因沉默引起冷调控基因PIF3等差异表达 Gene silencing induced differential expression of cold-regulated genes such as PIF3 | [ |
| 苜蓿 M. truncatula | 参与冷胁迫 Involved in cold stress | 构成lncRNA- MtCBFs调控网络 Construct lncRNA-MtCBFs regulatory network | [ | |
| 番茄 S. lycopersicum | 参与冷胁迫 Involved in cold stress | 竞争与共享miRNA来调控mRNA的表达 Competition and sharing of miRNAs to regulate mRNA expression | [ | |
| DE-lncRNAs | 甜椒 C. annuum | 参与冷胁迫 Involved in cold stress | 调控与冷损伤相关的靶基因 作为miRNAs前体响应冷胁迫 Regulating target genes related to cold damage Responding to cold stress as precursors of miRNAs | [ [ |
| 葡萄 V. vinifera | 参与冷胁迫 Involved in cold stress | 作为miRNAs的靶标 Being targets of miRNAs | [ | |
| 香蕉 M. balbisiana | 参与冷胁迫 Involved in cold stress | 调节类黄酮、蛋白激酶的生物合成以及TCA循环、硫传递系统等途径 Regulating the biosynthesis of flavonoids and protein kinases, as well as pathways such as the TCA cycle and sulfur transfer system | [ |
表1 植物中与低温相关的lncRNA
Table 1 Low temperature-related lncRNAs in plants
| lncRNA的名称 Name of lncRNA | 物种 Species | 功能 Function | 机制 Mechanism | 参考文献 Reference |
|---|---|---|---|---|
| COLDAIR | 拟南芥 A. thaliana | 参与春化作用 Involved in vernalization | 信号、引导分子,抑制FLC的表达 Signal, guide molecules, and inhibit the expression of FLC | [ |
| COLDWRAP | 拟南芥 A. thaliana | 参与春化作用 Involved in vernalization | 抑制FLC的表达 Inhibit the expression of FLC | [ |
| COOLAIR | 拟南芥 A. thaliana | 参与春化作用 Involved in vernalization | 增加H3K27me3和降低H3K36me3转录来抑制FLC Inhibition of FLC by increasing H3K27me3 and decreasing H3K36me3 transcription | [ |
| Svalka | 拟南芥 A. thaliana | 参与冷胁迫 Involved in cold stress | Svalka-asCBF1级联控制CBF1表达 Svalka-asCBF1 cascade controls CBF1 expression | [ |
| CIL1 | 拟南芥 A. thaliana | 参与冷胁迫 Involved in cold stress | 影响活性氧或渗透调节物质来响应冷胁迫 Responding to cold stress by affecting reactive oxygen species or osmotic regulators | [ |
| APOLO | 拟南芥 A. thaliana | 寒冷条件调节根毛伸长 Regulates root hair elongation under cold conditions | 与转录因子WRKY42相互作用 Interacting with the TF WRKY42 | [ |
| TE-lincRNA11195 | 拟南芥 A. thaliana | 参与冷胁迫 Involved in cold stress | 水杨酸刺激反应基因可能是潜在靶标 Salicylic acid-induced response genes may be potential targets | [ |
| DPA lncRNAs | 水稻 O. sativa | 参与冷胁迫 Involved in cold stress | 多聚腺苷酸化来响应冷胁迫 Polyadenylation in response to cold stress | [ |
| lncRNA-Chr03G0008 | 水稻 O. sativa | 参与冷胁迫 Involved in cold stress | 在幼苗中表达响应冷胁迫 Expressing response to cold stress in seedlings | [ |
| lncRNA-SVR | 水稻 O. sativa | 低温下与种子活力相关 Associated with seed viability under low temperature | 顺式基因SAUR家族成员相互作用 Interacting with the cis-gene family member SAUR | [ |
| lncR9A, lncR117, lncR616 | 东农冬麦1号 T. aestivum(Dn1) | 参与冷胁迫 Involved in cold stress | 竞争性结合miR398间接调节CSD1的表达 Indirectly regulates the expression of CSD1 by competitively binding miR398 | [ |
| Traes_2BS_7A04BF5D5 | 小麦 Durum wheat | 参与冷胁迫 Involved in cold stress | 以WCOR413冷驯化基因为靶标响应冷胁迫 Responding to cold stress targeting the cold acclimation gene WCOR413 | [ |
| CRR5 | 木薯 M. esculenta | 参与冷胁迫 Involved in cold stress | 与蛋白激酶基因协同表达响应低温胁迫 Co-expression with protein kinase gene in response to low temperature stress | [ |
| CRIR1 | 木薯 M. esculenta | 参与冷胁迫 Involved in cold stress | 招募MeCSP5来提高mRNA的翻译效率 Recruit MeCSP5 to improve the translation efficiency of mRNA | [ |
| XH123 | 棉花 G. hirsutum | 参与冷胁迫 Involved in cold stress | 基因沉默引起冷调控基因PIF3等差异表达 Gene silencing induced differential expression of cold-regulated genes such as PIF3 | [ |
| 苜蓿 M. truncatula | 参与冷胁迫 Involved in cold stress | 构成lncRNA- MtCBFs调控网络 Construct lncRNA-MtCBFs regulatory network | [ | |
| 番茄 S. lycopersicum | 参与冷胁迫 Involved in cold stress | 竞争与共享miRNA来调控mRNA的表达 Competition and sharing of miRNAs to regulate mRNA expression | [ | |
| DE-lncRNAs | 甜椒 C. annuum | 参与冷胁迫 Involved in cold stress | 调控与冷损伤相关的靶基因 作为miRNAs前体响应冷胁迫 Regulating target genes related to cold damage Responding to cold stress as precursors of miRNAs | [ [ |
| 葡萄 V. vinifera | 参与冷胁迫 Involved in cold stress | 作为miRNAs的靶标 Being targets of miRNAs | [ | |
| 香蕉 M. balbisiana | 参与冷胁迫 Involved in cold stress | 调节类黄酮、蛋白激酶的生物合成以及TCA循环、硫传递系统等途径 Regulating the biosynthesis of flavonoids and protein kinases, as well as pathways such as the TCA cycle and sulfur transfer system | [ |
| [1] | 谢虹, 杨兰, 李忠光. 脯氨酸在植物非生物胁迫耐性形成中的作用[J]. 生物技术通报, 2011(2): 23-27, 60. |
| Xie H, Yang L, Li ZG. The roles of proline in the formation of plant tolerance to abiotic stress[J]. Biotechnol Bull, 2011(2): 23-27, 60. | |
| [2] |
An D, Yang J, Zhang P. Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress[J]. BMC Genomics, 2012, 13(1): 64.
doi: 10.1186/1471-2164-13-64 |
| [3] | 录亚丹, 赵战胜, 张庭军. 北疆棉花苗期低温冷害的预防及补救措施[J]. 中国棉花, 2019, 46(6): 41-42. |
| Lu YD, Zhao ZS, Zhang TJ. Prevention and remedial measures of low temperature chilling injury of cotton seedlings in north Xinjiang[J]. China Cotton, 2019, 46(6): 41-42. | |
| [4] |
项洪涛, 郑殿峰, 何宁, 等. 植物对低温胁迫的生理响应及外源脱落酸缓解胁迫效应的研究进展[J]. 草业学报, 2021, 30(1): 208-219.
doi: 10.11686/cyxb2020091 |
| Xiang HT, Zheng DF, He N, et al. Research progress on the physiological response of plants to low temperature and the amelioration effcectiveness of exogenous ABA[J]. Acta Prataculturae Sin, 2021, 30(1): 208-219. | |
| [5] |
Zhou TJ, Zhang WX, Zhang LX, et al. 2021: A year of unprecedented climate extremes in eastern Asia, North America, and Europe[J]. Adv Atmos Sci, 2022, 39(10): 1598-1607.
doi: 10.1007/s00376-022-2063-9 |
| [6] |
Miquel M, James D Jr, Dooner H, et al. Arabidopsis requires polyunsaturated lipids for low-temperature survival[J]. Proc Natl Acad Sci Usa, 1993, 90(13): 6208-6212.
doi: 10.1073/pnas.90.13.6208 pmid: 11607410 |
| [7] |
Los DA. Membrane fluidity and temperature perception[J]. Plant Physiol, 1997, 115(3): 875-879.
pmid: 12223851 |
| [8] |
Alonso A, Queiroz CS, Magalhães AC. Chilling stress leads to increased cell membrane rigidity in roots of coffee(Coffea arabica L.) seedlings[J]. BBA Biomembr, 1997, 1323(1): 75-84.
doi: 10.1016/S0005-2736(96)00177-0 URL |
| [9] | 王兆. 低温胁迫对彩叶草的生理效应及抗寒性研究[D]. 福州: 福建农林大学, 2014. |
| Wang Z. Study on physiological effects and cold resistance of colored leaf grass under low temperature stress[D]. Fuzhou: Fujian Agriculture and Forestry University, 2014. | |
| [10] |
Li XY, Yang Y, Zhang LF, et al. Regulation on contents of endogenous hormones and Asr1 gene expression of maize seedling by exogenous ABA under low-temperature stress[J]. Acta Agron Sin, 2017, 43(1): 141.
doi: 10.3724/SP.J.1006.2017.00141 URL |
| [11] |
Nievola CC, Carvalho CP, Carvalho V, et al. Rapid responses of plants to temperature changes[J]. Temperature, 2017, 4(4): 371-405.
doi: 10.1080/23328940.2017.1377812 pmid: 29435478 |
| [12] |
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs[J]. Cell, 2009, 136(4): 629-641.
doi: 10.1016/j.cell.2009.02.006 pmid: 19239885 |
| [13] | 原佳沛, 张浩文, 鲁志. 新型长链非编码RNA(lncRNA)的生物信息学研究进展[J]. 生物化学与生物物理进展, 2013, 40(7): 634-640. |
| Yuan JP, Zhang HW, Lu Z. Progress on bioinformatic research of lncRNA[J]. Prog Biochem Biophys, 2013, 40(7): 634-640. | |
| [14] |
Waititu JK, Zhang CY, Liu J, et al. Plant non-coding RNAs: origin, biogenesis, mode of action and their roles in abiotic stress[J]. Int J Mol Sci, 2020, 21(21): 8401.
doi: 10.3390/ijms21218401 URL |
| [15] |
Nejat N, Mantri N. Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses[J]. Crit Rev Biotechnol, 2018, 38(1): 93-105.
doi: 10.1080/07388551.2017.1312270 pmid: 28423944 |
| [16] | 孙薏雯, 李金宝, 杨丹丹, 等. 长链非编码RNA在植物中的研究进展[J]. 山东农业大学学报: 自然科学版, 2020, 51(5): 968-974. |
| Sun YW, Li JB, Yang DD, et al. The research progress of long noncoding RNA in plants[J]. J Shandong Agric Univ Nat Sci Ed, 2020, 51(5): 968-974. | |
| [17] |
Wang JJ, Meng XW, Dobrovolskaya OB, et al. Non-coding RNAs and their roles in stress response in plants[J]. Genom Proteom Bioinform, 2017, 15(5): 301-312.
doi: 10.1016/j.gpb.2017.01.007 URL |
| [18] | 李睿, 罗云波. lncRNA及其生物学功能[J]. 农业生物技术学报, 2016, 24(4): 600-612. |
| Li R, Luo YB. LncRNA and its biological function[J]. Journal of Agricultural Biotechnology, 2016, 24(4): 600-612. | |
| [19] |
Hu HY, Wang MJ, Ding YH, et al. Transcriptomic repertoires depict the initiation of lint and fuzz fibres in cotton(Gossypium hirsutum L.)[J]. Plant Biotechnol J, 2018, 16(5): 1002-1012.
doi: 10.1111/pbi.2018.16.issue-5 URL |
| [20] | Salih IHI. 棉纤维发育起始和伸长阶段长链非编码RNA和mRNA的鉴定与表达[D]. 武汉: 华中农业大学, 2017. |
| Salih IHI. Identification and expression of long-chain non-coding RNA and mRNA in the initial and elongation stages of cotton fiber development[D]. Wuhan: Huazhong Agricultural University, 2017. | |
| [21] |
Wunderlich M, Groβ-Hardt R, Schöffl F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA[J]. Plant Mol Biol, 2014, 85(6): 541-550.
doi: 10.1007/s11103-014-0202-0 pmid: 24874772 |
| [22] | 刘琳营. 高温响应lncRNAs调控棉花花药育性的功能分析[D]. 武汉: 华中农业大学, 2021. |
| Liu LY. Functional analysis of high temperature response Lncrnas in regulating cotton anther fertility[D]. Wuhan: Huazhong Agricultural University, 2021. | |
| [23] |
Qin T, Zhao HY, Cui P, et al. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance[J]. Plant Physiol, 2017, 175(3): 1321-1336.
doi: 10.1104/pp.17.00574 pmid: 28887353 |
| [24] | 刘琳营, 苏晓俊, 闵玲. 植物中长链非编码RNA研究进展综述[J]. 江苏农业科学, 2021, 49(12): 12-19. |
| Liu LY, Su XJ, Min L. Review on research progress of long-chain non-coding RNA in plants[J]. Jiangsu Agric Sci, 2021, 49(12): 12-19. | |
| [25] | 李宁, 王柏柯, 王娟, 等. 植物长链非编码RNA的生物学功能研究进展[J]. 植物生理学报, 2019, 55(10): 1427-1435. |
| Li N, Wang BK, Wang J, et al. Advances in functional research of long non-coding RNAs in plants[J]. Plant Physiol J, 2019, 55(10): 1427-1435. | |
| [26] | 张爱晶, 郭兴玉, 何浩博, 等. 主要农作物lncRNA鉴定与分析研究进展[J]. 广东农业科学, 2020, 47(5): 1-10. |
| Zhang AJ, Guo XY, He HB, et al. Research progress on identification and analysis of lncRNA in major crops[J]. Guangdong Agric Sci, 2020, 47(5): 1-10. | |
| [27] |
Min L, Garbutt C, Tu CQ, et al. Potentials of long noncoding RNAs(LncRNAs)in sarcoma: From biomarkers to therapeutic targets[J]. Int J Mol Sci, 2017, 18(4): 731.
doi: 10.3390/ijms18040731 URL |
| [28] |
谭玉荣, 王丹, 高璇, 等. 植物长链非编码RNA研究进展[J]. 生物技术通报, 2018, 34(10): 1-10.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-0167 |
| Tan YR, Wang D, Gao X, et al. Research advance on plant long noncoding RNAs[J]. Biotechnol Bull, 2018, 34(10): 1-10. | |
| [29] | 艾雯, 李娇卓, 闵德栋, 等. 果蔬作物中长链非编码RNA研究进展[J]. 北方园艺, 2020(17): 124-130. |
| Ai W, Li JZ, Min DD, et al. Research progress of long noncoding RNAs on fruits and vegetables crops[J]. North Hortic, 2020(17): 124-130. | |
| [30] |
Rai MI, Alam M, Lightfoot DA, et al. Classification and experimental identification of plant long non-coding RNAs[J]. Genomics, 2019, 111(5): 997-1005.
doi: S0888-7543(18)30245-3 pmid: 29679643 |
| [31] |
Gao CX, Zheng XW, Li HB, et al. Roles of lncRNAs in rice: advances and challenges[J]. Rice Sci, 2020, 27(5): 384-395.
doi: 10.1016/j.rsci.2020.03.003 |
| [32] | Li S, Nayar S, Jia H, et al. The Arabidopsis hypoxia inducible AtR8 long non-coding RNA also contributes to plant defense and root elongation coordinating with WRKY genes under low levels of salicylic acid[J]. Noncoding RNA, 2020, 6(1): E8. |
| [33] |
Ding JH, Lu Q, Ouyang YD, et al. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice[J]. Proc Natl Acad Sci USA, 2012, 109(7): 2654-2659.
doi: 10.1073/pnas.1121374109 pmid: 22308482 |
| [34] |
Ariel F, Jegu T, Latrasse D, et al. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop[J]. Mol cell, 2014, 55(3): 383-396.
doi: 10.1016/j.molcel.2014.06.011 pmid: 25018019 |
| [35] |
Tian YK, Hou YK, Song Y. LncRNAs elevate plant adaptation under low temperature by maintaining local chromatin landscape[J]. Plant Signal Behav, 2022, 17(1): 2014677.
doi: 10.1080/15592324.2021.2014677 URL |
| [36] |
Campalans A, Kondorosi A, Crespi M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula[J]. Plant Cell, 2004, 16(4): 1047-1059.
pmid: 15037734 |
| [37] |
Wang Y, Luo XJ, Sun F, et al. Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice[J]. Nat Commun, 2018, 9(1): 3516.
doi: 10.1038/s41467-018-05829-7 pmid: 30158538 |
| [38] |
Jha UC, Nayyar H, Jha R, et al. Long non-coding RNAs: Emerging players regulating plant abiotic stress response and adaptation[J]. BMC Plant Biol, 2020, 20(1): 466.
doi: 10.1186/s12870-020-02595-x pmid: 33046001 |
| [39] |
Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity[J]. Nat Genet, 2007, 39(8): 1033-1037.
doi: 10.1038/ng2079 pmid: 17643101 |
| [40] | Fedak H, Palusinska M, Krzyczmonik K, et al. Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript[J]. Proc Natl Acad Sci USA, 2016, 113(48): E7846-E7855. |
| [41] |
Brockdorff N, Ashworth A, Kay GF, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome[J]. Nature, 1991, 351(6324): 329-331.
doi: 10.1038/351329a0 |
| [42] |
Brown CJ, Ballabio A, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome[J]. Nature, 1991, 349(6304): 38-44.
doi: 10.1038/349038a0 |
| [43] |
Lipshitz HD, Peattie DA, Hogness DS. Novel transcripts from the Ultrabithorax domain of the bithorax complex[J]. Genes Dev, 1987, 1(3): 307-322.
doi: 10.1101/gad.1.3.307 URL |
| [44] |
Moison M, Pacheco JM, Lucero L, et al. The lncRNA APOLO interacts with the transcription factor WRKY42 to trigger root hair cell expansion in response to cold[J]. Mol Plant, 2021, 14(6): 937-948.
doi: 10.1016/j.molp.2021.03.008 pmid: 33689931 |
| [45] | Liu WH, Cheng CZ, Lin YL, et al. Genome-wide identification and characterization of mRNAs and lncRNAs involved in cold stress in the wild banana(Musa itinerans)[J]. PLoS One, 2018, 13(7): e0200002. |
| [46] |
Jeon J, Kim J. Cold stress signaling networks in Arabidopsis[J]. J Plant Biol, 2013, 56(2): 69-76.
doi: 10.1007/s12374-013-0903-y URL |
| [47] |
Tang K, Zhao L, Ren Y, et al. The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes[J]. J Integr Plant Biol, 2020, 62(3): 258-263.
doi: 10.1111/jipb.v62.3 URL |
| [48] |
Kindgren P, Ard R, Ivanov M, et al. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation[J]. Nat Commun, 2018, 9(1): 4561.
doi: 10.1038/s41467-018-07010-6 pmid: 30385760 |
| [49] |
Liu GC, Liu FX, Wang Y, et al. A novel long noncoding RNA CIL1 enhances cold stress tolerance in Arabidopsis[J]. Plant Sci, 2022, 323: 111370.
doi: 10.1016/j.plantsci.2022.111370 URL |
| [50] |
Jampala P, Garhewal A, Lodha M. Functions of long non-coding RNA in Arabidopsis thaliana[J]. Plant Signal Behav, 2021, 16(9): 1925440.
doi: 10.1080/15592324.2021.1925440 URL |
| [51] |
Swiezewski S, Liu FQ, Magusin A, et al. Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target[J]. Nature, 2009, 462(7274): 799-802.
doi: 10.1038/nature08618 |
| [52] |
Ding YL, Shi YT, Yang SH. Molecular regulation of plant responses to environmental temperatures[J]. Mol plant, 2020, 13(4): 544-564.
doi: S1674-2052(20)30034-4 pmid: 32068158 |
| [53] |
Severing E, Faino L, Jamge S, et al. Arabidopsis thaliana ambient temperature responsive lncRNAs[J]. BMC Plant Biol, 2018, 18(1): 145.
doi: 10.1186/s12870-018-1362-x pmid: 30005624 |
| [54] |
Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA[J]. Science, 2011, 331(6013): 76-79.
doi: 10.1126/science.1197349 pmid: 21127216 |
| [55] |
Kim DH, Sung S. Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs[J]. Dev Cell, 2017, 40(3): 302-312.
doi: 10.1016/j.devcel.2016.12.021 URL |
| [56] | Tian Y, Zheng H, Zhang F, et al. PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR[J]. Sci Adv, 2019, 5(4): eaau7246. |
| [57] |
Csorba T, Questa JI, Sun QW, et al. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization[J]. Proc Natl Acad Sci USA, 2014, 111(45): 16160-16165.
doi: 10.1073/pnas.1419030111 pmid: 25349421 |
| [58] |
Pan YH, Liang HF, Gao LJ, et al. Transcriptomic profiling of germinating seeds under cold stress and characterization of the cold-tolerant gene LTG5 in rice[J]. BMC Plant Biol, 2020, 20(1): 371.
doi: 10.1186/s12870-020-02569-z |
| [59] |
Yuan JP, Li JR, Yang Y, et al. Stress-responsive regulation of long non-coding RNA polyadenylation in Oryza sativa[J]. Plant J, 2018, 93(5): 814-827.
doi: 10.1111/tpj.2018.93.issue-5 URL |
| [60] |
Shin SY, Jeong JS, Lim JY, et al. Transcriptomic analyses of rice(Oryza sativa)genes and non-coding RNAs under nitrogen starvation using multiple omics technologies[J]. BMC genomics, 2018, 19(1): 532.
doi: 10.1186/s12864-018-4897-1 |
| [61] |
Gao QL, Liu JZ, Weng HB, et al. A long noncoding RNA derived from lncRNA-mRNA networks modulates seed vigor[J]. Int J Mol Sci, 2022, 23(16): 9472.
doi: 10.3390/ijms23169472 URL |
| [62] | Leng Y, Sun J, Wang JG, et al. Genome-wide lncRNAs identification and association analysis for cold-responsive genes at the booting stage in rice(Oryza sativa L.)[J]. Plant Genome, 2020, 13(2): e20020. |
| [63] | 卢秋巍. 冬小麦抗寒lncRNA筛选及与tae-miR398应答低温胁迫的互作研究[D]. 哈尔滨: 东北农业大学, 2018. |
| Lu QW. Screening of cold-resistant lncRNA in winter wheat and its interaction with tae-miR398 in response to low temperature stress[D]. Harbin:Northeast Agricultural University, 2018. | |
| [64] |
Díaz ML, Soresi DS, Basualdo J, et al. Transcriptomic response of durum wheat to cold stress at reproductive stage[J]. Mol Biol Rep, 2019, 46(2): 2427-2445.
doi: 10.1007/s11033-019-04704-y pmid: 30798485 |
| [65] |
Lu QW, Guo FY, Xu QH, et al. LncRNA improves cold resistance of winter wheat by interacting with miR398[J]. Funct Plant Biol, 2020, 47(6): 544-557.
doi: 10.1071/FP19267 pmid: 32345432 |
| [66] |
Suksamran R, Saithong T, Thammarongtham C, et al. Genomic and transcriptomic analysis identified novel putative cassava lncRNAs involved in cold and drought stress[J]. Genes, 2020, 11(4): 366.
doi: 10.3390/genes11040366 URL |
| [67] | 沈婕, 董世满, 李淑霞, 等. 木薯中lncRNA-CRR5在低温胁迫下的功能分析[J]. 热带作物学报, 2021, 42(1): 40-46. |
| Shen J, Dong SM, Li SX, et al. Identification of lncRNA-CRR5 in response to low temperature stress in cassava[J]. Chin J Trop Crops, 2021, 42(1): 40-46. | |
| [68] |
Li S, Cheng Z, Dong S, et al. Global identification of full-length cassava lncRNAs unveils the role of cold-responsive intergenic lncRNA 1 in cold stress response[J]. Plant Cell Environ, 2022, 45(2): 412-426.
doi: 10.1111/pce.v45.2 URL |
| [69] | 李玉强. 低温冷害对尉犁县棉花生长发育的影响分析[J]. 农业灾害研究, 2018, 8(5): 42-43. |
| Li YQ. Effect of chilling damage on growth and development of cotton in Yuli County[J]. J Agric Catastrophol, 2018, 8(5): 42-43. | |
| [70] | 曹泽毅. 棉花冷害相关长链非编码RNA的筛选与功能分析[D]. 杭州: 浙江大学, 2018. |
| Cao ZY. Screening and functional analysis of long-chain non-coding RNA related to cotton chilling injury[D]. Hangzhou: Zhejiang University, 2018. | |
| [71] |
Liu ZY, Li XL, Li F, et al. Mechanisms underlying the effects of fall dormancy on the cold acclimation and winter hard-iness of Medicago sativa[J]. Chin J Plant Ecol, 2015, 39(6): 635-648.
doi: 10.17521/cjpe.2015.0061 URL |
| [72] |
Zhao MG, Wang TZ, Sun TY, et al. Identification of tissue-specific and cold-responsive lncRNAs in Medicago truncatula by high-throughput RNA sequencing[J]. BMC Plant Biol, 2020, 20(1): 99.
doi: 10.1186/s12870-020-2301-1 |
| [73] | Zhao DY, Shen L, Fan B, et al. Physiological and genetic properties of tomato fruits from 2 cultivars differing in chilling tolerance at cold storage[J]. J Food Sci, 2009, 74(5): C348-C352. |
| [74] |
Wang YX, Gao LP, Zhu BZ, et al. Integrative analysis of long non-coding RNA acting as ceRNAs involved in chilling injury in tomato fruit[J]. Gene, 2018, 667: 25-33.
doi: S0378-1119(18)30513-4 pmid: 29753809 |
| [75] |
Zuo JH, Wang YX, Zhu BZ, et al. Analysis of the coding and non-coding RNA transcriptomes in response to bell pepper chilling[J]. Int J Mol Sci, 2018, 19(7): 2001.
doi: 10.3390/ijms19072001 URL |
| [76] |
Baruah PM, Krishnatreya DB, Bordoloi KS, et al. Genome wide identification and characterization of abiotic stress responsive lncRNAs in Capsicum annuum[J]. Plant Physiol Biochem, 2021, 162: 221-236.
doi: 10.1016/j.plaphy.2021.02.031 URL |
| [77] |
Wang P, Dai L, Ai J, et al. Identification and functional prediction of cold-related long non-coding RNA(lncRNA)in grapevine[J]. Sci Rep, 2019, 9(1): 1-15.
doi: 10.1038/s41598-018-37186-2 |
| [1] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [2] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [3] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [4] | 徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219. |
| [5] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [6] | 郭怡婷, 赵文菊, 任延靖, 赵孟良. 菊芋NAC转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(6): 217-232. |
| [7] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [8] | 王兵, 赵会纳, 余婧, 余世洲, 雷波. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22. |
| [9] | 史建磊, 宰文珊, 苏世闻, 付存念, 熊自立. 番茄青枯病抗性相关miRNA的鉴定与表达分析[J]. 生物技术通报, 2023, 39(5): 233-242. |
| [10] | 张新博, 崔浩亮, 史佩华, 高锦春, 赵顺然, 陶晨雨. 低起始量的免疫共沉淀技术研究进展[J]. 生物技术通报, 2023, 39(4): 227-235. |
| [11] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [12] | 葛颜锐, 赵冉, 徐静, 李若凡, 胡云涛, 李瑞丽. 植物维管形成层发育及其调控的研究进展[J]. 生物技术通报, 2023, 39(3): 13-25. |
| [13] | 蒋路园, 丰美静, 杜雨晴, 邸葆, 陈段芬, 邱德有, 杨艳芳. 红豆杉低温半致死温度和低温胁迫下紫杉烷含量[J]. 生物技术通报, 2023, 39(3): 232-242. |
| [14] | 刘铖霞, 孙宗艳, 罗云波, 朱鸿亮, 曲桂芹. bHLH转录因子的磷酸化调控植物生理功能的研究进展[J]. 生物技术通报, 2023, 39(3): 26-34. |
| [15] | 赵孟良, 郭怡婷, 任延靖. 菊芋WRKY转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(2): 116-125. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||