








生物技术通报 ›› 2024, Vol. 40 ›› Issue (5): 269-279.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1032
蒋文萍1,2( ), 冉秋萍1,2, 刘家书1,2, 张慧敏1,2, 张迪1,2, 江正兵1,2, 李华南1,2(
), 冉秋萍1,2, 刘家书1,2, 张慧敏1,2, 张迪1,2, 江正兵1,2, 李华南1,2( )
)
收稿日期:2023-11-02
出版日期:2024-05-26
发布日期:2024-06-13
通讯作者:
李华南,女,博士,副教授,研究方向:生物催化与转化;E-mail: huananli@hubu.edu.cn作者简介:蒋文萍,女,硕士研究生,研究方向:生物催化与转化;E-mail: 1828269638@qq.com
基金资助:
JIANG Wen-ping1,2( ), RAN Qiu-ping1,2, LIU Jia-shu1,2, ZHANG Hui-min1,2, ZHANG Di1,2, JIANG Zheng-bing1,2, LI Hua-nan1,2(
), RAN Qiu-ping1,2, LIU Jia-shu1,2, ZHANG Hui-min1,2, ZHANG Di1,2, JIANG Zheng-bing1,2, LI Hua-nan1,2( )
)
Received:2023-11-02
Published:2024-05-26
Online:2024-06-13
摘要:
【目的】旨在探究不同来源碳水化合物结合域(CBM)对山毛榉木聚糖的结合能力,并将具有较高结合能力的外源CBM融合到链霉菌L10904木聚糖酶(XYN)的C端和N端,以探究外源CBM对木聚糖酶酶学性质的影响。【方法】通过底物吸附方法,利用考马斯亮蓝G250法检测溶液中CBM在吸附前后的浓度,计算CBM的底物结合率,筛选到了结合木聚糖能力较好的CBM1和CBM4。为了探究对底物结合能力高的CBM融合位置对木聚糖酶酶学性质的影响,将CBM1和CBM4通过柔性连接肽分别与XYN的C端和N端融合,并在大肠杆菌中表达获得4种重组酶,分别命名为CBM1-XYN、XYN-CBM1、CBM4-XYN和XYN-CBM4。【结果】CBM1和CBM4与木聚糖结合率分别为89%和95%。在60℃, pH 7.0反应条件下,XYN、CBM1-XYN、XYN-CBM1、CBM4-XYN和XYN-CBM4的比活力分别是32 274.81、49 342.21、602.48、230.42和2 362.24 U/mg,CBM1-XYN比活力较XYN比活力提高了1.5倍。酶学性质分析表明,CBM1使XYN温度稳定性和pH稳定性得到了提高,将XYN和CBM1-XYN分别在60℃孵育1 h,CBM1-XYN残余酶活力和XYN残余酶活力分别为81%和28%;在pH 3-11范围内,CBM1-XYN在4℃孵育12 h后能够保持90%以上的酶活力。【结论】在大肠杆菌中成功异源表达了链霉菌来源的木聚糖酶,筛选到了对底物结合率高的两种CBM1和CBM4,并通过蛋白质融合技术成功将CBM融合到XYN上,获得酶学性质得到改良的CBM1-XYN,能够提高木聚糖酶的温度稳定性、pH耐受性及比酶活。
蒋文萍, 冉秋萍, 刘家书, 张慧敏, 张迪, 江正兵, 李华南. 碳水化合物结合域对木聚糖酶酶学性质的影响[J]. 生物技术通报, 2024, 40(5): 269-279.
JIANG Wen-ping, RAN Qiu-ping, LIU Jia-shu, ZHANG Hui-min, ZHANG Di, JIANG Zheng-bing, LI Hua-nan. Effects of Carbohydrate-binding Modules on the Enzymatic Properties of Xylanase[J]. Biotechnology Bulletin, 2024, 40(5): 269-279.
| 代号Code name | 名称 Name | 生物信息数据库GenBank | 来源 Source | ||
|---|---|---|---|---|---|
| 菌属 Genus | 酶 Enzyme | ||||
| 1 | CBM1 1号 | BAD01163.1 | Trametes hirsuta | Endoglucanase | |
| 2 | CBM1 2号 | AAF35251.1 | T. versicolor | Cellobiohydrolase | |
| 3 | CBM1 3号 | CAM98445.1 | Acremonium thermophilum | Cellulose 1,4-beta-cellobiosidase | |
| 4 | CBM1 4号 | CAA83846.1 | Trichoderma reesei | Endo-1,4-beta-glucanase V | |
| 5 | CBM1 5号 | CAA37878.1 | T. viride | Cellobiohydrolase | |
| 6 | CBM1 6号 | AAQ21383.1 | T. viride | Endoglucanase III | |
| 7 | CBM1 7号 | AAQ76092.1 | T. viride | Eellobiohydrolase I | |
| 8 | CBM1 8号 | AAQ76094.1 | T. viride | Cellobiohydrolase II | |
| 1# | CBM4 | AF039030.1 | Hungateiclostridium thermocellum JW20 | Cellulose 1,4-beta-cellobiosidase | |
| 2# | CBM3 1号 | ABN54273.1 | H. thermocellum ATCC 27405 | Cellulosome anchoring protein cohesin region | |
| 3# | CBM3 2号 | ABN51281.1 | H. thermocellum ATCC 27405 | Glycoside hydrolase family 9 | |
| 4# | CBM2 | AAB42115.1 | Thermobifida fusca YX | Beta-1,4-endoglucanase precursor | |
表1 不同类型CBM相关介绍
Table 1 Introduction to different types of CBM
| 代号Code name | 名称 Name | 生物信息数据库GenBank | 来源 Source | ||
|---|---|---|---|---|---|
| 菌属 Genus | 酶 Enzyme | ||||
| 1 | CBM1 1号 | BAD01163.1 | Trametes hirsuta | Endoglucanase | |
| 2 | CBM1 2号 | AAF35251.1 | T. versicolor | Cellobiohydrolase | |
| 3 | CBM1 3号 | CAM98445.1 | Acremonium thermophilum | Cellulose 1,4-beta-cellobiosidase | |
| 4 | CBM1 4号 | CAA83846.1 | Trichoderma reesei | Endo-1,4-beta-glucanase V | |
| 5 | CBM1 5号 | CAA37878.1 | T. viride | Cellobiohydrolase | |
| 6 | CBM1 6号 | AAQ21383.1 | T. viride | Endoglucanase III | |
| 7 | CBM1 7号 | AAQ76092.1 | T. viride | Eellobiohydrolase I | |
| 8 | CBM1 8号 | AAQ76094.1 | T. viride | Cellobiohydrolase II | |
| 1# | CBM4 | AF039030.1 | Hungateiclostridium thermocellum JW20 | Cellulose 1,4-beta-cellobiosidase | |
| 2# | CBM3 1号 | ABN54273.1 | H. thermocellum ATCC 27405 | Cellulosome anchoring protein cohesin region | |
| 3# | CBM3 2号 | ABN51281.1 | H. thermocellum ATCC 27405 | Glycoside hydrolase family 9 | |
| 4# | CBM2 | AAB42115.1 | Thermobifida fusca YX | Beta-1,4-endoglucanase precursor | |
| 引物 Primers | 序列 Sequence(5'-3') |
|---|---|
| pET23a-F | gctagcatgactggtggacagcaaatgggt |
| pET23a-R | gtatatctccttcttaaagttaaacaaaattatttctagagggaaaccgtt |
| sfgfp-sumo-F | aactttaagaaggagatatacatgcatcatcaccatcaccatatggtgagcaagggc |
| sfgfp-sumo-R1 | ctgccgcttaccgtcctgctgtccaccaatctgttctctgtgagc |
| sfgfp-sumo-R2 | accgcactggccccagacggctccaccaatctgttctctgtg |
| sfgfp-sumo-R3 | tctttcatacaaaaggtcgtTtccaccaatctgttctctgtgagcctc |
| xyn-F | gctcacagagaacagattggtggacagcaggacggtaagcggcag |
| xyn-R | tttgctgtccaccagtcatgctagccatcagccgctgaccgtgatgttcga |
| cbm1-xyn-F | cacagagaacagattggtggagccgtctggggccagtgcggt |
| cbm1-xyn-R | gctgtccaccagtcatgctagctcagccgctgaccgtgat |
| cbm4-xyn-F | gaggctcacagagaacagattggtggaAacgaccttttgtatgaaaga |
| cbm4-xyn-R | tccaccagtcatgctagctcagccgctgaccgtgatgttcga |
| xyn-cbm1-R | tttgctgtccaccagtcatgctagcctggcactgcgagtagt |
| xyn-cbm4-R | tttgctgtccaccagtcatgctagcaggatcgtagagagatacatcatcaagg |
| cbm1-F | ggcggtggtgggtcgggtggcggtggctcggccgtctggggccag |
| cbm1-R | accgcactggccccagacggctccaccaatctgttctctgtg |
| cbm4-F | ggcggtggtgggtcgggtggcggtggctcgAacgaccttttgtat |
| cbm4-R | cgacccaccaccgcccgagccaccgccaccaggatcgtagagagatac |
表2 构建融合基因的引物序列
Table 2 Primer sequences for constructing fusion genes
| 引物 Primers | 序列 Sequence(5'-3') |
|---|---|
| pET23a-F | gctagcatgactggtggacagcaaatgggt |
| pET23a-R | gtatatctccttcttaaagttaaacaaaattatttctagagggaaaccgtt |
| sfgfp-sumo-F | aactttaagaaggagatatacatgcatcatcaccatcaccatatggtgagcaagggc |
| sfgfp-sumo-R1 | ctgccgcttaccgtcctgctgtccaccaatctgttctctgtgagc |
| sfgfp-sumo-R2 | accgcactggccccagacggctccaccaatctgttctctgtg |
| sfgfp-sumo-R3 | tctttcatacaaaaggtcgtTtccaccaatctgttctctgtgagcctc |
| xyn-F | gctcacagagaacagattggtggacagcaggacggtaagcggcag |
| xyn-R | tttgctgtccaccagtcatgctagccatcagccgctgaccgtgatgttcga |
| cbm1-xyn-F | cacagagaacagattggtggagccgtctggggccagtgcggt |
| cbm1-xyn-R | gctgtccaccagtcatgctagctcagccgctgaccgtgat |
| cbm4-xyn-F | gaggctcacagagaacagattggtggaAacgaccttttgtatgaaaga |
| cbm4-xyn-R | tccaccagtcatgctagctcagccgctgaccgtgatgttcga |
| xyn-cbm1-R | tttgctgtccaccagtcatgctagcctggcactgcgagtagt |
| xyn-cbm4-R | tttgctgtccaccagtcatgctagcaggatcgtagagagatacatcatcaagg |
| cbm1-F | ggcggtggtgggtcgggtggcggtggctcggccgtctggggccag |
| cbm1-R | accgcactggccccagacggctccaccaatctgttctctgtg |
| cbm4-F | ggcggtggtgggtcgggtggcggtggctcgAacgaccttttgtat |
| cbm4-R | cgacccaccaccgcccgagccaccgccaccaggatcgtagagagatac |

图1 不同种类CBM的SDS-PAGE分析及其对山毛榉木聚糖的吸附能力 A:SDS-PAGE分析纯蛋白(M:蛋白分子标准;1-8:CBM1家族;1#:CBM4;2#:CBM3家族1号;3#:CBM3家族2号;4#:CBM2);B:不同种类CBM对山毛榉木聚糖吸附能力;图中误差线表示标准偏差,下同
Fig. 1 SDS-PAGE analysis of different types of CBM and adsorption capacity on beech xylan A: SDS-PAGE analysis of pure protein(M: Protein molecular standards; 1-8: family CBM1; 1#: CBM4; 2#: CBM3 family No. 1; 3#: CBM3 family No. 2; 4#: CBM2). B: Adsorption capacity of different types of CBM on beech xylan. The error line in the figure indicates the standard deviation. The same below

图2 融合酶SDS-PAGE分析 A:粗酶溶液SDS-PAGE分析(M:Marker;1:SFGFP-SUMO-XYN;2:SFGFP-SUMO-CBM1-XYN;3:SFGFP-SUMO-XYN-CBM1;4:SFGFP-SUMO-CBM4-XYN;5:SFGFP-SUMO-XYN-CBM4);B:纯酶SDS-PAGE分析(M:Marker;1:SFGFP-SUMO-XYN;2:SFGFP-SUMO-CBM1-XYN;3:SFGFP-SUMO-XYN-CBM1);C:ULP1酶作用于SUMO,切割融合蛋白,并对SFGFP-SUMO和靶蛋白进行SDS-PAGE分析(M:Marker;1:SFGFP-SUMO & XYN:2:SFGFP-SUMO & CBM1-XYN;3:SFGFP-SUMO & XYN-CBM1)
Fig. 2 SDS-PAGE analysis of fusion enzyme A: SDS-PAGE analysis of crude supernatant enzyme solution. B: Pure enzyme SDS-PAGE analysis. C: ULP1 enzyme acts on SUMO, cleaves fusion protein, and SDS-PAGE analysis of SFGFP-SUMO and target protein
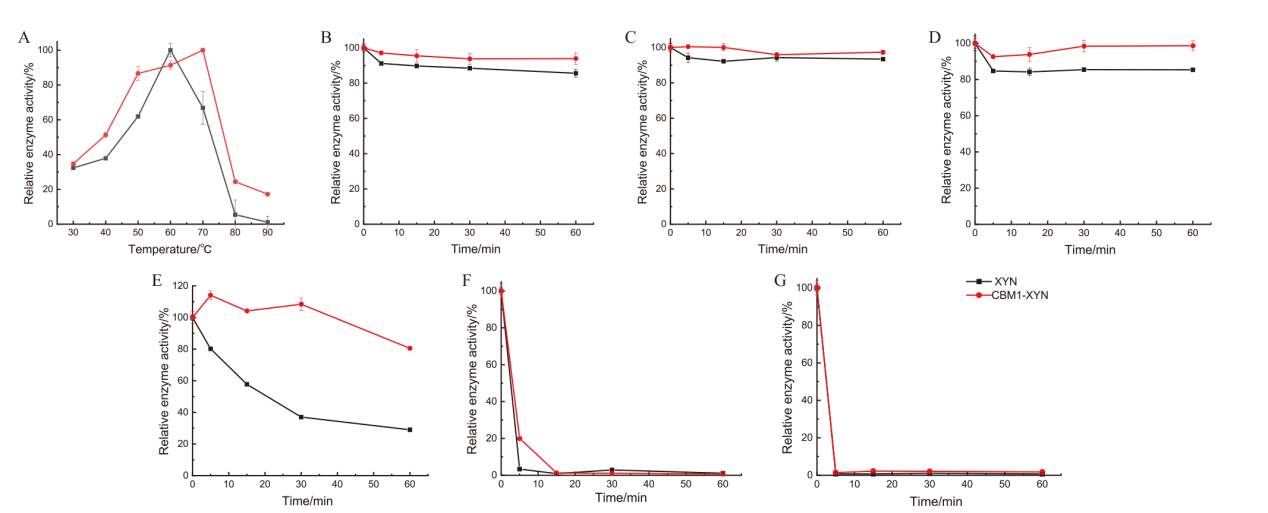
图4 XYN和CBM1-XYN的最适反应温度(A)和温度稳定性(B-G)
Fig. 4 Optimal temperature (A) and temperature stability (B-G) for XYN and CBM1-XYN B: 30℃; C: 40℃; D: 50℃; E: 60℃; F: 70℃; G: 80℃
| 金属离子和化学试剂 Metal ion and chemical reagent | 相对酶活Relative enzyme activity/% | ||
|---|---|---|---|
| XYN | CBM1-XYN | ||
| FeCl3 | 92.6±1.67 | 101.9±5.07 | |
| NaCl | 95.4±4.14 | 109.6±3.29 | |
| CaCl2 | 93.4±1.84 | 106.8±7.74 | |
| MnSO4 | 73.1±1.89 | 92.6±1.22 | |
| KCl | 101.7±4.14 | 104.9±2.85 | |
| FeSO4 | 75.3±0.66 | 81±2.11 | |
| CoCl2 | 98.8±1.71 | 101.4±1.13 | |
| MgCl2 | 94.8±2.20 | 108.7±1.66 | |
| (NH4)2SO4 | 98.4±1.45 | 101±1.02 | |
表3 金属离子及化学试剂对酶活力影响
Table 3 Effect of metal ions and chemical reagents on enzyme activity
| 金属离子和化学试剂 Metal ion and chemical reagent | 相对酶活Relative enzyme activity/% | ||
|---|---|---|---|
| XYN | CBM1-XYN | ||
| FeCl3 | 92.6±1.67 | 101.9±5.07 | |
| NaCl | 95.4±4.14 | 109.6±3.29 | |
| CaCl2 | 93.4±1.84 | 106.8±7.74 | |
| MnSO4 | 73.1±1.89 | 92.6±1.22 | |
| KCl | 101.7±4.14 | 104.9±2.85 | |
| FeSO4 | 75.3±0.66 | 81±2.11 | |
| CoCl2 | 98.8±1.71 | 101.4±1.13 | |
| MgCl2 | 94.8±2.20 | 108.7±1.66 | |
| (NH4)2SO4 | 98.4±1.45 | 101±1.02 | |
| 酶 Enzyme | Vmax /(mg·mL-1· min-1) | Km /(mg· mL-1) | Kcat/Km /(mL· mg-1·min-1) |
|---|---|---|---|
| XYN | 1.564 7 | 0.012 5 | 335.879 9 |
| CBM1-XYN | 2.842 5 | 0.009 5 | 891.909 0 |
表4 XYN和CBM1-XYN的动力学参数
Table 4 Dynamics parameters of XYN and CBM1-XYN
| 酶 Enzyme | Vmax /(mg·mL-1· min-1) | Km /(mg· mL-1) | Kcat/Km /(mL· mg-1·min-1) |
|---|---|---|---|
| XYN | 1.564 7 | 0.012 5 | 335.879 9 |
| CBM1-XYN | 2.842 5 | 0.009 5 | 891.909 0 |
| [1] |
Zoghlami A, Paës G. Lignocellulosic biomass: understanding recalcitrance and predicting hydrolysis[J]. Front Chem, 2019, 7: 874.
doi: 10.3389/fchem.2019.00874 pmid: 31921787 |
| [2] | Madhavan A, Arun KB, Sindhu R, et al. Design and genome engineering of microbial cell factories for efficient conversion of lignocellulose to fuel[J]. Bioresour Technol, 2023, 370: 128555. |
| [3] | Liu YJ, Wang J, Bao CL, et al. Characterization of a novel GH10 xylanase with a carbohydrate binding module from Aspergillus sulphureus and its synergistic hydrolysis activity with cellulase[J]. Int J Biol Macromol, 2021, 182: 701-711. |
| [4] |
Nguyen STC, Freund HL, Kasanjian J, et al. Function, distribution, and annotation of characterized cellulases, xylanases, and chitinases from CAZy[J]. Appl Microbiol Biotechnol, 2018, 102(4): 1629-1637.
doi: 10.1007/s00253-018-8778-y pmid: 29359269 |
| [5] |
Moreira LRS, Filho EXF. Insights into the mechanism of enzymatic hydrolysis of xylan[J]. Appl Microbiol Biotechnol, 2016, 100(12): 5205-5214.
doi: 10.1007/s00253-016-7555-z pmid: 27112349 |
| [6] | Rahmani N, Kahar P, Lisdiyanti P, et al. GH-10 and GH-11 Endo-1, 4-β-xylanase enzymes from Kitasatospora sp. produce xylose and xylooligosaccharides from sugarcane bagasse with no xylose inhibition[J]. Bioresour Technol, 2019, 272: 315-325. |
| [7] | You S, Li J, Zhang F, et al. Loop engineering of a thermostable GH10 xylanase to improve low-temperature catalytic performance for better synergistic biomass-degrading abilities[J]. Bioresour Technol, 2021, 342: 125962. |
| [8] | Joshi JB, Priyadharshini R, Uthandi S. Glycosyl hydrolase 11(xynA)gene with xylanase activity from thermophilic bacteria isolated from thermal springs[J]. Microb Cell Fact, 2022, 21(1): 62. |
| [9] | Xiong K, Yan ZX, Liu JY, et al. Inter domain interactions influence the substrate affinity and hydrolysis product specificity of xylanase from Streptomyces chartreusis L1105[J]. Ann Microbiol, 2020, 70(1): 1-12. |
| [10] | Li JF, Wang CJ, Hu D, et al. Engineering a family 27 carbohydrate-binding module into an Aspergillus usamii β-mannanase to perfect its enzymatic properties[J]. J Biosci Bioeng, 2017, 123(3): 294-299. |
| [11] |
Boraston AB, Revett TJ, Boraston CM, et al. Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate binding module, TmCBM27[J]. Structure, 2003, 11(6): 665-675.
pmid: 12791255 |
| [12] | Liu LW, Zeng LY, Wang SY, et al. Activity and thermostability increase of xylanase following transplantation with modules sub-divided from hyper-thermophilic CBM9_1-2[J]. Process Biochem, 2012, 47(5): 853-857. |
| [13] | Wang HL, Qi XH, Gao S, et al. Biochemical characterization of an engineered bifunctional xylanase/feruloyl esterase and its synergistic effects with cellulase on lignocellulose hydrolysis[J]. Bioresour Technol, 2022, 355: 127244. |
| [14] | Li XQ, Xia JL, Zhu XY, et al. Construction and characterization of bifunctional cellulases: Caldicellulosiruptor-sourced endoglucanase, CBM, and exoglucanase for efficient degradation of lignocellulose[J]. Biochem Eng J, 2019, 151: 107363. |
| [15] | Shi QC, Abdel-Hamid AM, Sun ZY, et al. Carbohydrate-binding modules facilitate the enzymatic hydrolysis of lignocellulosic biomass: releasing reducing sugars and dissociative lignin available for producing biofuels and chemicals[J]. Biotechnol Adv, 2023, 65: 108126. |
| [16] | Mandal A, Thakur A, Goyal A. Role of carbohydrate binding modules, CBM3A and CBM3B in stability and catalysis by a β-1, 4 endoglucanase, AtGH9C-CBM3A-CBM3B from Acetivibrio thermocellus ATCC 27405[J]. Int J Biol Macromol, 2023, 242(Pt 4): 125164. |
| [17] | Zhou JL, Harindintwali JD, Yang WH, et al. Engineering of a chitosanase fused to a carbohydrate-binding module for continuous production of desirable chitooligosaccharides[J]. Carbohydr Polym, 2021, 273: 118609. |
| [18] | Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database(CAZy)in 2013[J]. Nucleic Acids Res, 2014, 42(Database issue): D490-D495. |
| [19] | Kang DH, You SK, Joo YC, et al. Synergistic effect of the enzyme complexes comprising agarase, carrageenase and neoagarobiose hydrolase on degradation of the red algae[J]. Bioresour Technol, 2018, 250: 666-672. |
| [20] |
Chalak A, Villares A, Moreau C, et al. Influence of the carbohydrate-binding module on the activity of a fungal AA9 lytic polysaccharide monooxygenase on cellulosic substrates[J]. Biotechnol Biofuels, 2019, 12: 206.
doi: 10.1186/s13068-019-1548-y pmid: 31508147 |
| [21] | Liu T, Zhang Y, Lu XM, et al. Binding affinity of family 4 carbohydrate binding module on cellulose films of nanocrystals and nanofibrils[J]. Carbohydr Polym, 2021, 251: 116725. |
| [22] | Lee JP, Shin ES, Cho MY, et al. Roles of carbohydrate-binding module(CBM)of an endo-β-1, 4-glucanase(Cel5L)from Bacillus sp. KD1014 in thermostability and small-substrate hydrolyzing activity[J]. J Microbiol Biotechnol, 2018, 28(12): 2036-2045. |
| [23] | Thongekkaew J, Ikeda H, Masaki K, et al. Fusion of cellulose binding domain from Trichoderma reesei CBHI to Cryptococcus sp. S-2 cellulase enhances its binding affinity and its cellulolytic activity to insoluble cellulosic substrates[J]. Enzyme Microb Technol, 2013, 52(4/5): 241-246. |
| [24] | Pan RH, Hu YM, Long LK, et al. Extra carbohydrate binding module contributes to the processivity and catalytic activity of a non-modular hydrolase family 5 endoglucanase from Fomitiporia mediterranea MF3/22[J]. Enzyme Microb Technol, 2016, 91: 42-51. |
| [25] | Hu YM, Li HN, Ran QP, et al. Effect of carbohydrate binding modules alterations on catalytic activity of glycoside hydrolase family 6 exoglucanase from Chaetomium thermophilum to cellulose[J]. Int J Biol Macromol, 2021, 191: 222-229. |
| [26] | Tajwar R, Shahid S, Zafar R, et al. Impact of orientation of carbohydrate binding modules family 22 and 6 on the catalytic activity of Thermotoga maritima xylanase XynB[J]. Enzyme Microb Technol, 2017, 106: 75-82. |
| [27] | Rooijakkers BJM, Arola S, Velagapudi R, et al. Different effects of carbohydrate binding modules on the viscoelasticity of nanocellulose gels[J]. Biochem Biophys Rep, 2020, 22: 100766. |
| [28] | Li H, Lu ZJ, Hao MS, et al. Family 92 carbohydrate-binding modules specific for β-1, 6-glucans increase the thermostability of a bacterial chitinase[J]. Biochimie, 2023, 212: 153-160. |
| [29] | Kurniati A, Puspaningsih NNT, Putri KDA, et al. Heterologous fusion gene expression and characterization of a novel carbohydrate binding module(Cbm36)to laccase(Lcc2)[J]. Biocatal Agric Biotechnol, 2022, 42: 102377. |
| [30] | Hoffmam ZB, Zanphorlin LM, Cota J, et al. Xylan-specific carbohydrate-binding module belonging to family 6 enhances the catalytic performance of a GH11 endo-xylanase[J]. N Biotechnol, 2016, 33(4): 467-472. |
| [31] | Khan MIM, Sajjad M, Sadaf S, et al. The nature of the carbohydrate binding module determines the catalytic efficiency of xylanase Z of Clostridium thermocellum[J]. J Biotechnol, 2013, 168(4): 403-408. |
| [32] |
Yang A, Cheng JS, Liu M, et al. Sandwich fusion of CBM9_2 to enhance xylanase thermostability and activity[J]. Int J Biol Macromol, 2018, 117: 586-591.
doi: S0141-8130(18)30844-4 pmid: 29852224 |
| [33] | Xu WX, Liu YJ, Ye YX, et al. C-Terminal carbohydrate-binding module 9_2 fused to the N-terminus of GH11 xylanase from Aspergillus niger[J]. Biotechnol Lett, 2016, 38(10): 1739-1745. |
| [34] |
Li Q, Sun BG, Jia HY, et al. Engineering a xylanase from Streptomyce rochei L10904 by mutation to improve its catalytic characteristics[J]. Int J Biol Macromol, 2017, 101: 366-372.
doi: S0141-8130(17)30498-1 pmid: 28356235 |
| [35] | Wu QH, Zhang CN, Zhu WJ, et al. Improved thermostability, acid tolerance as well as catalytic efficiency of Streptomyces rameus L2001 GH11 xylanase by N-terminal replacement[J]. Enzyme Microb Technol, 2023, 162: 110143. |
| [36] |
Han YJ, Gao PX, Yu WG, et al. Thermostability enhancement of chitosanase CsnA by fusion a family 5 carbohydrate-binding module[J]. Biotechnol Lett, 2017, 39(12): 1895-1901.
doi: 10.1007/s10529-017-2406-2 pmid: 28748352 |
| [37] | Sari B, Faiz O, Genc B, et al. New xylanolytic enzyme from Geobacillus galactosidasius BS61 from a geothermal resource in Turkey[J]. Int J Biol Macromol, 2018, 119: 1017-1026. |
| [38] | van Dyk JS, Sakka M, Sakka K, et al. Characterisation of the multi-enzyme complex xylanase activity from Bacillus licheniformis SVD1[J]. Enzyme Microb Technol, 2010, 47(4): 174-177. |
| [39] |
Hong J, Ye XH, Zhang YH. Quantitative determination of cellulose accessibility to cellulase based on adsorption of a nonhydrolytic fusion protein containing CBM and GFP with its applications[J]. Langmuir, 2007, 23(25): 12535-12540.
doi: 10.1021/la7025686 pmid: 17988165 |
| [40] | He HB, Liu JJ, Wang YT, et al. Site-directed mutagenesis of family GH10 Aspergillus fumigatus xylanase A and the interaction with Oryza sativa xylanase inhibitor protein[J]. Biocatal Agric Biotechnol, 2023, 54: 102920. |
| [41] | Hong SJ, Park BR, Lee HN, et al. Carbohydrate-binding module of cycloisomaltooligosaccharide glucanotransferase from Thermoanaerobacter thermocopriae improves its cyclodextran production[J]. Enzyme Microb Technol, 2022, 157: 110023. |
| [1] | 杨伟杰, 杨周林, 朱浩东, 魏煜, 刘君, 刘训. 地衣素合成酶关键模块 LchAD 蛋白的性质和功能研究[J]. 生物技术通报, 2024, 40(3): 322-332. |
| [2] | 杨佳慧, 孙玉萍, 陆雅宁, 刘欢, 卢存福, 陈玉珍. 拟南芥AtTERT对大肠杆菌非生物胁迫抗性的影响[J]. 生物技术通报, 2022, 38(2): 1-9. |
| [3] | 李志豪, 张鸽, 貊志杰, 邓帅军, 李佳轶, 张海波, 刘晓晖, 刘好宝. 一株产木聚糖酶的蜡样芽孢杆菌对雪茄烟叶成分及发酵产物的影响[J]. 生物技术通报, 2022, 38(2): 105-112. |
| [4] | 吴蓉, 曹佳睿, 曹君, 刘飞翔, 杨猛, 苏二正. 南极假丝酵母脂肪酶B基因在大肠杆菌中的表达和发酵优化[J]. 生物技术通报, 2021, 37(2): 138-148. |
| [5] | 余琴, 马现永, 邓盾, 王永飞. 海氏肠球菌IDO5对猪粪废水中吲哚降解条件优化及降解途径分析[J]. 生物技术通报, 2021, 37(12): 113-123. |
| [6] | 黄坤龙, 苏小运, 姚斌. 生长抑素和耐热木聚糖酶的融合表达及性质研究[J]. 生物技术通报, 2020, 36(9): 235-243. |
| [7] | 杨彬, 李小波, 周林, 区佩渝, 金小宝. 同步分泌高效纤维素酶和木聚糖酶菌株YB的鉴定及其酶学性质研究[J]. 生物技术通报, 2020, 36(2): 110-118. |
| [8] | 李鹏昊, 梁严予, 王彦伟, 关洋, 逄文强, 田克恭. 非洲猪瘟病毒K196R和A240L蛋白的可溶性表达及酶活力分析[J]. 生物技术通报, 2020, 36(11): 70-75. |
| [9] | 肖政, 范里, 谭文松. 半乳糖流加对CHO细胞生长代谢及其表达的Fc融合蛋白糖基化的影响[J]. 生物技术通报, 2019, 35(8): 138-145. |
| [10] | 邓晓芬, 杨晓佳, 易天红, 冯英, 柯潇, 赖维莉. 融合蛋白基因与抗体基因电转染CHO-S细胞的条件摸索优化[J]. 生物技术通报, 2019, 35(4): 223-228. |
| [11] | 陈欣宁, 赵亮, 范里, 刘旭平, 谭文松. 氨浓度升高对CHO细胞维持期抗体融合蛋白的表达及N-糖基化的影响[J]. 生物技术通报, 2019, 35(3): 93-102. |
| [12] | 张伊丽, 刘照平, 陆敏依, 蔡明勇, 吴根鹏. CO2分压对贴壁细胞MRC-5连续传代培养的影响[J]. 生物技术通报, 2019, 35(10): 169-173. |
| [13] | 顾源, 寻子琦, 郑菲, 涂涛, 姚斌, 罗会颖. 来源于嗜热真菌Talaromyces leycettanus JCM12802 的类膨胀素基因鉴定及功能探究[J]. 生物技术通报, 2018, 34(10): 172-181. |
| [14] | 陈春野,刘剑,朱瑞,李姝璇,叶江辉,王玮,潘德全,徐飞海,程通,夏宁邵. 大肠杆菌可溶性表达人鳞状上皮细胞癌抗原的制备及应用[J]. 生物技术通报, 2017, 33(9): 252-258. |
| [15] | 王亚美,魏原杰,艾新宇,刘小宁. 棉蚜His-CYP6J1融合蛋白的分离纯化及多克隆抗体的制备[J]. 生物技术通报, 2017, 33(5): 164-169. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||