








生物技术通报 ›› 2024, Vol. 40 ›› Issue (4): 297-305.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1068
收稿日期:2023-11-14
出版日期:2024-04-26
发布日期:2024-04-30
通讯作者:
王彩虹同为本文通讯作者作者简介:王彩虹,女,博士,助理研究员,研究方向:肿瘤、微生物; E-mail: 13520137104@163.com
基金资助:
WANG Cai-hong( ), JIANG Meng-yuan, SHAO Yu-han
), JIANG Meng-yuan, SHAO Yu-han
Received:2023-11-14
Published:2024-04-26
Online:2024-04-30
摘要:
【目的】探讨VI型分泌系统(type VI secretion system, T6SS)新的效应蛋白及其功能。【方法】以铜绿假单胞菌PAO1为研究对象,通过基因敲除、免疫印迹、免疫共沉淀、种内和种间竞争、大肠杆菌毒性实验以及结晶紫生物膜等实验探索PA0423的分泌方式和功能。【结果】免疫印迹和免疫共沉淀实验表明PA0423可以与PA0262相互作用,且PA0423的分泌依赖于H2-T6SS;竞争实验表明PA0423是H2-T6SS依赖的抗菌效应蛋白,且具有种间竞争优势;PA0423具有大肠杆菌毒性且PA0422为其免疫蛋白;PA0423能够影响细菌生物膜的形成。【结论】PA0423为H2-T6SS依赖的抗菌效应蛋白,其具有杀菌功能且影响细菌生物膜的形成。
王彩虹, 蒋梦媛, 邵煜涵. 铜绿假单胞菌潜在VI型分泌系统效应蛋白PA0423的鉴定及功能研究[J]. 生物技术通报, 2024, 40(4): 297-305.
WANG Cai-hong, JIANG Meng-yuan, SHAO Yu-han. Identification and Functional Study of T6SS Effector Protein PA0423 of Pseudomonas aeruginosa[J]. Biotechnology Bulletin, 2024, 40(4): 297-305.
| 菌株 Strain | 特点 Characteristic | 来源 Source |
|---|---|---|
| DH5a | 大肠杆菌基因克隆菌株 | 全式金 |
| BL21(DE3)pLysS | 大肠杆菌基因表达菌株 | 全式金 |
| PAO1 | 野生型 | 本实验室 |
| S17-1 | 接合菌株 | 本实验室 |
| PAO1△rets | PAO1的rets敲除突变 | 本实验室 |
| PAO1△rets△clpV1 | PAO1的rets和clpV1敲除突变 | 本实验室 |
| PAO1△rets△clpV2 | PAO1的rets和clpV2敲除突变 | 本实验室 |
| PAO1△rets△clpV3 | PAO1的rets和clpV3敲除突变 | 本实验室 |
| PAO1△PA0262 | PAO1的PA0262敲除突变 | 本实验室 |
| PAO1△PA0423 | PAO1的PA0423敲除突变 | 本实验室 |
| PAO1△PA0423+PME-PA0423 | 包含PME6032-PA0423重组质粒的PAO1的PA0423敲除菌 | 本实验室 |
| PAO1△PA0422-0423 | PAO1的PA0422和PA0423敲除突变 | 本实验室 |
表1 本研究所用的菌株
Table 1 Strains used in this study
| 菌株 Strain | 特点 Characteristic | 来源 Source |
|---|---|---|
| DH5a | 大肠杆菌基因克隆菌株 | 全式金 |
| BL21(DE3)pLysS | 大肠杆菌基因表达菌株 | 全式金 |
| PAO1 | 野生型 | 本实验室 |
| S17-1 | 接合菌株 | 本实验室 |
| PAO1△rets | PAO1的rets敲除突变 | 本实验室 |
| PAO1△rets△clpV1 | PAO1的rets和clpV1敲除突变 | 本实验室 |
| PAO1△rets△clpV2 | PAO1的rets和clpV2敲除突变 | 本实验室 |
| PAO1△rets△clpV3 | PAO1的rets和clpV3敲除突变 | 本实验室 |
| PAO1△PA0262 | PAO1的PA0262敲除突变 | 本实验室 |
| PAO1△PA0423 | PAO1的PA0423敲除突变 | 本实验室 |
| PAO1△PA0423+PME-PA0423 | 包含PME6032-PA0423重组质粒的PAO1的PA0423敲除菌 | 本实验室 |
| PAO1△PA0422-0423 | PAO1的PA0422和PA0423敲除突变 | 本实验室 |
| 引物 Primer | 序列 Sequence | 作用 Function |
|---|---|---|
| PA0423-up-F | ACAGCTATGACATGATTAC GAATTCGCGGGTCGCCTTCGCCGGCG AGCAC | 构建pk18-PA0423 |
| PA0423-up-R | GCGTTTTTCGTTTCAGGTATCCGCGGACATTCTCCTTAACGTGGTTGTTG | 构建pk18-PA0423 |
| PA0423-down-F | CAACAACCACGTTAAGGAGAATGTCCGCGGATACCTGAAACGAAAAACGC | 构建pk18-PA0423 |
| PA0423-down-R | CTGCAGGTCGACTCTAGA GGATCCGGCGCTTGAGGATGATGCCGTTCAC | 构建pk18-PA0423 |
| PA0422-0423-up-F | ACAGCTATGACATGATTAC GAATTCGGTGCTGGCGGAAGCCTTCG | 构建pk18-PA0422-0423 |
| PA0422-0423-up-R | TTCGTTTCAGGTATCCGCGAAAAACTTCCTTGAAAAGC | 构建pk18-PA0422-0423 |
| PA0422-0423-down-F | GCTTTTCAAGGAAGTTTTTCGCGGATACCTGAAACGAA | 构建pk18-PA0422-0423 |
| PA0422-0423-down-R | CTGCAGGTCGACTCTAGA GGATCCTGCCGTTCACCTGGGGACGC | 构建pk18-PA0422-0423 |
| PME-0423-F | ATTTCACACAGGAAACA GAATTCATGCTGAAGAAGACCCTTGC | 构建PME6032-PA0423-FLAG |
| PME-0423-flag-R | ACTGATCCGCTAGTCCGAGGCCTCGAGCTTATCGTCGTCATCCTTGTAAT C | 构建PME6032-PA0423-FLAG |
| PME-flag-R | ACTGATCCGCTAGTCCGAGGC CTCGAGCTTATCGTCGTCATCCTTGTAAT C | 构建PME6032-PA0423-FLAG |
| RSF-PA0423-F | CATCACCACAGCCAGGATCC GAATTCGATGCTGAAGAAGACCCTTGCCG | 构建RSFduet-PA0423-myc |
| RSF-PA0423-R | GTCGAAGGCATTCGCCAGGAGCAGAAACTCATCTCTGAAGAGGATCTG AAGCTTGCGGCCGCATAATGCT | 构建RSFduet-PA0423-myc |
| pET-PA0262-F | CATATGGCAGATCTCAATTG GATATCATGCGTCAAA GGGACCTGAA ATTC | 构建pETduet-PA0262 |
| pET-PA0262-R | GCAGCGGTTTCTTTACCAGA CTCGAGGTATCCCGTTGGGAAGTTTTTCAG | 构建pETduet-PA0262 |
| pET28-PA0423-F | ATGCTGAAGAAGACCCTTGC | 构建pET28-PA423 |
| pET28-PA0423-R | CTTATCGTCGTCATCCTTG | 构建pET28-PA423 |
表2 本研究所用引物
Table 2 Primers used in this study
| 引物 Primer | 序列 Sequence | 作用 Function |
|---|---|---|
| PA0423-up-F | ACAGCTATGACATGATTAC GAATTCGCGGGTCGCCTTCGCCGGCG AGCAC | 构建pk18-PA0423 |
| PA0423-up-R | GCGTTTTTCGTTTCAGGTATCCGCGGACATTCTCCTTAACGTGGTTGTTG | 构建pk18-PA0423 |
| PA0423-down-F | CAACAACCACGTTAAGGAGAATGTCCGCGGATACCTGAAACGAAAAACGC | 构建pk18-PA0423 |
| PA0423-down-R | CTGCAGGTCGACTCTAGA GGATCCGGCGCTTGAGGATGATGCCGTTCAC | 构建pk18-PA0423 |
| PA0422-0423-up-F | ACAGCTATGACATGATTAC GAATTCGGTGCTGGCGGAAGCCTTCG | 构建pk18-PA0422-0423 |
| PA0422-0423-up-R | TTCGTTTCAGGTATCCGCGAAAAACTTCCTTGAAAAGC | 构建pk18-PA0422-0423 |
| PA0422-0423-down-F | GCTTTTCAAGGAAGTTTTTCGCGGATACCTGAAACGAA | 构建pk18-PA0422-0423 |
| PA0422-0423-down-R | CTGCAGGTCGACTCTAGA GGATCCTGCCGTTCACCTGGGGACGC | 构建pk18-PA0422-0423 |
| PME-0423-F | ATTTCACACAGGAAACA GAATTCATGCTGAAGAAGACCCTTGC | 构建PME6032-PA0423-FLAG |
| PME-0423-flag-R | ACTGATCCGCTAGTCCGAGGCCTCGAGCTTATCGTCGTCATCCTTGTAAT C | 构建PME6032-PA0423-FLAG |
| PME-flag-R | ACTGATCCGCTAGTCCGAGGC CTCGAGCTTATCGTCGTCATCCTTGTAAT C | 构建PME6032-PA0423-FLAG |
| RSF-PA0423-F | CATCACCACAGCCAGGATCC GAATTCGATGCTGAAGAAGACCCTTGCCG | 构建RSFduet-PA0423-myc |
| RSF-PA0423-R | GTCGAAGGCATTCGCCAGGAGCAGAAACTCATCTCTGAAGAGGATCTG AAGCTTGCGGCCGCATAATGCT | 构建RSFduet-PA0423-myc |
| pET-PA0262-F | CATATGGCAGATCTCAATTG GATATCATGCGTCAAA GGGACCTGAA ATTC | 构建pETduet-PA0262 |
| pET-PA0262-R | GCAGCGGTTTCTTTACCAGA CTCGAGGTATCCCGTTGGGAAGTTTTTCAG | 构建pETduet-PA0262 |
| pET28-PA0423-F | ATGCTGAAGAAGACCCTTGC | 构建pET28-PA423 |
| pET28-PA0423-R | CTTATCGTCGTCATCCTTG | 构建pET28-PA423 |
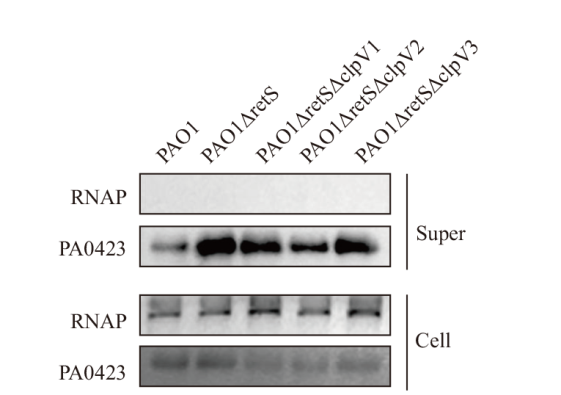
图1 PA0423的分泌检测及验证 免疫印迹检测PA0423在野生型PAO1以及不同PAO1突变细菌中的表达水平以及细胞上清中的分泌情况,RNAP被用作内参
Fig. 1 Detection and validation of PA0423 secretion Expression and secretion levels of PA0423 in wild-type and different mutant PAO1 bacteria were detected by Western blotting. RNAP was used as an internal control
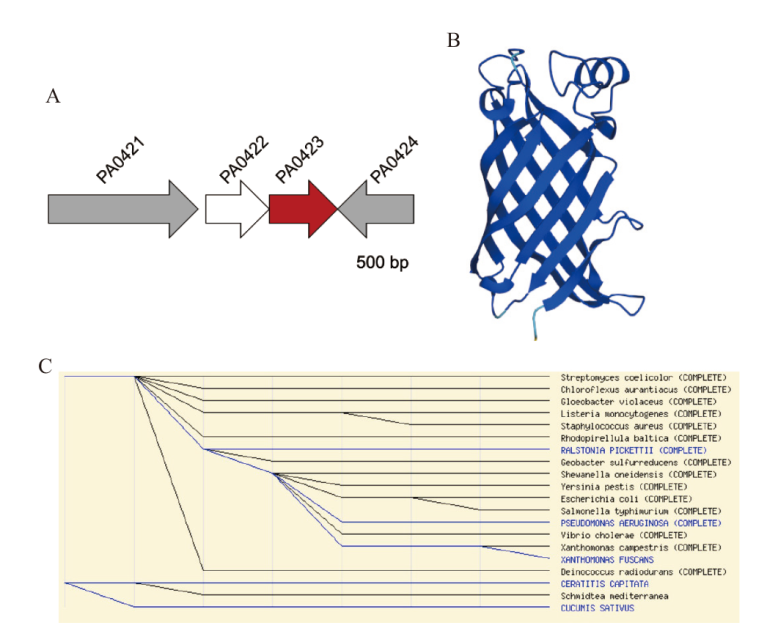
图3 PA0423的蛋白结构和分布情况 a:PA0423的基因排列图;b:AlphaFold预测PA0423的结构模型(AF-Q9I690-F1);c:MEROPS数据库分析小蛋白酶S85家族的分布
Fig. 3 Protein structure and distribution of PA0423 a: Gene arrangement map of PA0423. b: Structural model of PA0423(AF-Q9I690-F1)by AlphaFold. c: Distribution of small protease S85 family analyzed by MEROPS database
| 分析 Analysis | 编号 Accession No. | 描述 Description | Interpro 编号 Interpro accession | Interpro 描述 Interpro description | 氨基酸起始Amino acid start | 氨基酸终止 Amino acid stop | 误差 E-value |
|---|---|---|---|---|---|---|---|
| Hamap | MF_00780 | Protein YceI | IPR023480 | Uncharacterised protein family UPF0312/YceI | 2 | 191 | 45.339172 |
| PANTHER | PTHR34406 | Protein YCEI | - | - | 2 | 190 | 6.6E-62 |
| SUPERFAMILY | SSF101874 | YceI-like | IPR036761 | Lipid/polyisoprenoid-binding,Yce-like superfamily | 22 | 191 | 2.88E-56 |
| Gene3D | G3DSA:2.40128.110 | - | IPR036761 | Lipid/polyisoprenoid-binding,Yce-like superfamily | 24 | 191 | 1.7E-58 |
| FunFam | G3DSA:2.40128.110:FF:000001 | UPF0312 protein HMPREF3014_17255 | - | - | 24 | 191 | 3.7E-118 |
| SMART | SM00867 | YceI_2 | IPR007372 | Lipid/polyisoprenoid-binding,Yce-like | 25 | 189 | 1.3E-71 |
| Pfam | PF04264 | YceI-like domain | IPR007372 | Lipid/polyisoprenoid-binding,Yce-like | 26 | 187 | 2.9E-41 |
表3 通过 Interpro 数据库进行功能预测
Table 3 Function prediction via Interpro database
| 分析 Analysis | 编号 Accession No. | 描述 Description | Interpro 编号 Interpro accession | Interpro 描述 Interpro description | 氨基酸起始Amino acid start | 氨基酸终止 Amino acid stop | 误差 E-value |
|---|---|---|---|---|---|---|---|
| Hamap | MF_00780 | Protein YceI | IPR023480 | Uncharacterised protein family UPF0312/YceI | 2 | 191 | 45.339172 |
| PANTHER | PTHR34406 | Protein YCEI | - | - | 2 | 190 | 6.6E-62 |
| SUPERFAMILY | SSF101874 | YceI-like | IPR036761 | Lipid/polyisoprenoid-binding,Yce-like superfamily | 22 | 191 | 2.88E-56 |
| Gene3D | G3DSA:2.40128.110 | - | IPR036761 | Lipid/polyisoprenoid-binding,Yce-like superfamily | 24 | 191 | 1.7E-58 |
| FunFam | G3DSA:2.40128.110:FF:000001 | UPF0312 protein HMPREF3014_17255 | - | - | 24 | 191 | 3.7E-118 |
| SMART | SM00867 | YceI_2 | IPR007372 | Lipid/polyisoprenoid-binding,Yce-like | 25 | 189 | 1.3E-71 |
| Pfam | PF04264 | YceI-like domain | IPR007372 | Lipid/polyisoprenoid-binding,Yce-like | 26 | 187 | 2.9E-41 |
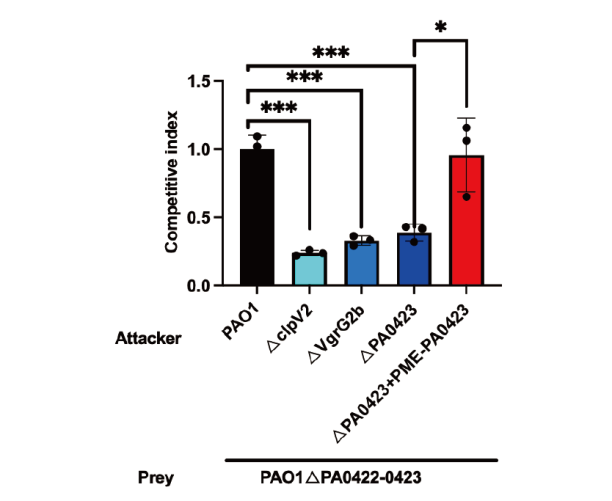
图4 PA0423是铜绿假单胞菌H2-T6SS依赖的抗菌效应蛋白 PAO1ΔPA0422-0423菌株与各攻击菌株在37℃下竞争24 h的种内竞争试验。值和误差线用SD ±平均值(n= 3 个生物学重复)。采用Dunnett检验的单因素方差分析,*P <0.05;**P <0.01;***P<0.001,下同
Fig. 4 PA0423 is a H2-T6SS-dependent antibacterial effector protein of P. aeruginosa The PAO1ΔPA0422-0423 strain competed with the attacker strains at 37℃ for 24 h. Values and error bars were SD ± average(n= 3 biological replicates). One-way ANOVA using Dunnett’s test, *P <0.05, **P <0.01, and ***P<0.001; the same below
| [1] |
Rossi E, La Rosa R, Bartell JA, et al. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis[J]. Nat Rev Microbiol, 2021, 19(5): 331-342.
doi: 10.1038/s41579-020-00477-5 pmid: 33214718 |
| [2] |
Silby MW, Winstanley C, Godfrey SAC, et al. Pseudomonas genomes: diverse and adaptable[J]. FEMS Microbiol Rev, 2011, 35(4): 652-680.
doi: 10.1111/j.1574-6976.2011.00269.x pmid: 21361996 |
| [3] |
Yasmin L, Jansson AL, Panahandeh T, et al. Delineation of exoenzyme S residues that mediate the interaction with 14-3-3 and its biological activity[J]. FEBS J, 2006, 273(3): 638-646.
pmid: 16420486 |
| [4] |
Goldberg JB, Pier GB. The role of the CFTR in susceptibility to Pseudomonas aeruginosa infections in cystic fibrosis[J]. Trends Microbiol, 2000, 8(11): 514-520.
pmid: 11121762 |
| [5] |
Scheetz MH, Hoffman M, Bolon MK, et al. Morbidity associated with Pseudomonas aeruginosa bloodstream infections[J]. Diagn Microbiol Infect Dis, 2009, 64(3): 311-319.
doi: 10.1016/j.diagmicrobio.2009.02.006 URL |
| [6] |
Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system[J]. Cell Host Microbe, 2014, 15(1): 9-21.
doi: 10.1016/j.chom.2013.11.008 pmid: 24332978 |
| [7] | Ziemski M, Leodolter J, Taylor G, et al. Genome-wide interaction screen for Mycobacterium tuberculosis ClpCP protease reveals toxin-antitoxin systems as a major substrate class[J]. FEBS J, 2021, 288(1): 111-126. |
| [8] | Chen LH, Zou YR, Kronfl AA, et al. Type VI secretion system of Pseudomonas aeruginosa is associated with biofilm formation but not environmental adaptation[J]. MicrobiologyOpen, 2020, 9(3): e991. |
| [9] | Wang TT, Du X, Ji LX, et al. Pseudomonas aeruginosa T6SS-mediated molybdate transport contributes to bacterial competition during anaerobiosis[J]. Cell Rep, 2021, 35(2): 108957. |
| [10] | Hernandez RE, Gallegos-Monterrosa R, Coulthurst SJ. Type VI secretion system effector proteins: effective weapons for bacterial competitiveness[J]. Cell Microbiol, 2020, 22(9): e13241. |
| [11] |
Pissaridou P, Allsopp LP, Wettstadt S, et al. The Pseudomonas aeruginosa T6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors[J]. Proc Natl Acad Sci USA, 2018, 115(49): 12519-12524.
doi: 10.1073/pnas.1814181115 pmid: 30455305 |
| [12] |
Ma JL, Pan ZH, Huang JH, et al. The Hcp proteins fused with diverse extended-toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems[J]. Virulence, 2017, 8(7): 1189-1202.
doi: 10.1080/21505594.2017.1279374 pmid: 28060574 |
| [13] | Sana TG, Berni B, Bleves S. The T6SSs of Pseudomonas aeruginosa strain PAO1 and their effectors: beyond bacterial-cell targeting[J]. Front Cell Infect Microbiol, 2016, 6: 61. |
| [14] | Han YY, Wang TT, Chen GK, et al. A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition[J]. PLoS Pathog, 2019, 15(12): e1008198. |
| [15] |
Mougous JD, Cuff ME, Raunser S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus[J]. Science, 2006, 312(5779): 1526-1530.
doi: 10.1126/science.1128393 pmid: 16763151 |
| [16] |
Jurėnas D, Journet L. Activity, delivery, and diversity of Type VI secretion effectors[J]. Mol Microbiol, 2021, 115(3): 383-394.
doi: 10.1111/mmi.14648 pmid: 33217073 |
| [17] |
Hood RD, Singh P, Hsu F, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria[J]. Cell Host Microbe, 2010, 7(1): 25-37.
doi: 10.1016/j.chom.2009.12.007 URL |
| [18] | Yang XB, Long MX, Shen XH. Effector-Immunity pairs provide the T6SS nanomachine its offensive and defensive capabilities[J]. Molecules, 2018, 23(5): 1009. |
| [19] |
Russell AB, Hood RD, Bui NK, et al. Type VI secretion delivers bacteriolytic effectors to target cells[J]. Nature, 2011, 475(7356): 343-347.
doi: 10.1038/nature10244 |
| [20] | Pérez-Lorente AI, Molina-Santiago C, de Vicente A, et al. Sporulation activated via σW protects Bacillus from a Tse1 peptidoglycan hydrolase type VI secretion system effector[J]. Microbiol Spectr, 2023, 11(2): e0504522. |
| [21] |
Coulthurst S. The Type VI secretion system: a versatile bacterial weapon[J]. Microbiology, 2019, 165(5): 503-515.
doi: 10.1099/mic.0.000789 |
| [22] | Sana TG, Baumann C, Merdes A, et al. Internalization of Pseudomonas aeruginosa strain PAO1 into epithelial cells is promoted by interaction of a T6SS effector with the microtubule network[J]. mBio, 2015, 6(3): e00712. |
| [23] |
Rumbaugh KP, Sauer K. Biofilm dispersion[J]. Nat Rev Microbiol, 2020, 18(10): 571-586.
doi: 10.1038/s41579-020-0385-0 pmid: 32533131 |
| [24] |
Ju XY, Chen J, Zhou MX, et al. Combating Pseudomonas aeruginosa biofilms by a chitosan-PEG-peptide conjugate via changes in assembled structure[J]. ACS Appl Mater Interfaces, 2020, 12(12): 13731-13738.
doi: 10.1021/acsami.0c02034 URL |
| [25] | de Sousa T, Hébraud M, Dapkevicius MLNE, et al. Genomic and metabolic characteristics of the pathogenicity in Pseudomonas aeruginosa[J]. Int J Mol Sci, 2021, 22(23): 12892. |
| [26] | Lin JS, Cheng JL, Chen KQ, et al. The icmF3 locus is involved in multiple adaptation- and virulence-related characteristics in Pseudomonas aeruginosa PAO1[J]. Front Cell Infect Microbiol, 2015, 5: 70. |
| [27] |
Tang AH, Caballero AR, Marquart ME, et al. Mechanism of Pseudomonas aeruginosa small protease(PASP), a corneal virulence factor[J]. Invest Ophthalmol Vis Sci, 2018, 59(15): 5993-6002.
doi: 10.1167/iovs.18-25834 URL |
| [28] |
Tang AH, Marquart ME, Fratkin JD, et al. Properties of PASP: a Pseudomonas protease capable of mediating corneal erosions[J]. Invest Ophthalmol Vis Sci, 2009, 50(8): 3794-3801.
doi: 10.1167/iovs.08-3107 URL |
| [29] |
Handa N, Terada T, Doi-Katayama Y, et al. Crystal structure of a novel polyisoprenoid-binding protein from Thermus thermophilus HB8[J]. Protein Sci, 2005, 14(4): 1004-1010.
doi: 10.1110/ps.041183305 URL |
| [30] |
Sisinni L, Cendron L, Favaro G, et al. Helicobacter pylori acidic stress response factor HP1286 is a YceI homolog with new binding specificity[J]. FEBS J, 2010, 277(8): 1896-1905.
doi: 10.1111/ejb.2010.277.issue-8 URL |
| [1] | 刘星雨, 李洁, 朱龙佼, 李相阳, 许文涛. 铜绿假单胞菌适配体的获得及应用[J]. 生物技术通报, 2024, 40(1): 186-193. |
| [2] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [3] | 扈丽丽, 林柏荣, 王宏洪, 陈建松, 廖金铃, 卓侃. 最短尾短体线虫转录组及潜在效应蛋白分析[J]. 生物技术通报, 2023, 39(3): 254-266. |
| [4] | 贺丽娜, 冯源, 石慧敏, 叶建仁. 具有杀线活性马尾松内生细菌的筛选与鉴定[J]. 生物技术通报, 2022, 38(8): 159-166. |
| [5] | 胡珊, 梁卫驱, 黄皓, 徐匆, 罗华建, 胡楚维, 黄晓彦, 陈仕丽. 中药渣堆肥中解磷细菌的筛选、鉴定及其拮抗作用[J]. 生物技术通报, 2022, 38(3): 92-102. |
| [6] | 洪军, 卫夏怡, 吉冰洁, 叶延欣, 程天赐. 铜绿假单胞菌对鲎素耐药前后的差异表达基因及SNP变化研究[J]. 生物技术通报, 2021, 37(9): 191-202. |
| [7] | 邓苗苗, 郭晓黎. 植物响应寄生线虫侵染机制的研究进展[J]. 生物技术通报, 2021, 37(7): 25-34. |
| [8] | 彭焕, 赵薇, 姚珂, 蒋陈, 黄文坤, 孔令安, 郑经武, 彭德良. 植物寄生线虫基因组学研究进展[J]. 生物技术通报, 2021, 37(7): 3-13. |
| [9] | 程英, 靳明辉, 萧玉涛. 鳞翅目昆虫基因编辑技术研究进展[J]. 生物技术通报, 2020, 36(3): 18-28. |
| [10] | 朱平, 杜力杰, 孟昆, 薛娟, 杨瑾, 李姗. 三型分泌系统效应蛋白调控细胞凋亡和焦亡的研究进展[J]. 生物技术通报, 2019, 35(4): 178-187. |
| [11] | 张婧波,吴剑荣,蒋芸,杨迪,詹晓北. 以抽油烟机废油为原料发酵产鼠李糖脂的研究[J]. 生物技术通报, 2018, 34(5): 195-200. |
| [12] | 张帆, 李树垚, 张子豪, 蔡雪凤, 任岩, 刘凯, 侯翠艳, 易欣欣, 高秀芝. 铜绿假单胞菌检测方法的比较与优化[J]. 生物技术通报, 2018, 34(3): 67-74. |
| [13] | 王洪洋, 秦丽娟, 唐唯, 田振东. 致病疫霉RXLR效应蛋白相关研究进展[J]. 生物技术通报, 2018, 34(2): 102-111. |
| [14] | 朱慧明, 张彦, 杨洪江. 高产铁载体假单胞菌的筛选及其对铁氧化物的利用[J]. 生物技术通报, 2015, 31(9): 177-182. |
| [15] | 陈春琳, 刘祥, 王成祥, 张晓娟, 俱雄, 张涛, 吴三桥. 铜绿假单胞菌外膜蛋白I的原核表达、纯化及免疫保护作用研究[J]. 生物技术通报, 2015, 31(7): 207-213. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||